6CBD
 
 | |
6JOU
 
 | | Crystal structure of the human nucleosome containing H2A.Z.1 S42R | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Mizukami, Y, Kurumizaka, H. | | Deposit date: | 2019-03-23 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-based design of an H2A.Z.1 mutant stabilizing a nucleosome in vitro and in vivo.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
7PMN
 
 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation II) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha-binding protein, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
6AQR
 
 | | SAGA DUB module Ubp8(C146A)/Sgf11/Sus1/Sgf73 bound to monoubiquitin | | Descriptor: | Polyubiquitin-C, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Morrow, M.E, Morgan, M.T, Wolberger, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins.
EMBO Rep., 19, 2018
|
|
5OF3
 
 | |
1H6K
 
 | | nuclear Cap Binding Complex | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, CBP80 | | Authors: | Mazza, C, Ohno, M, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Nuclear CAP Binding Complex
Mol.Cell, 8, 2001
|
|
3EVE
 
 | |
3EVF
 
 | | Crystal structure of Me7-GpppA complex of yellow fever virus methyltransferase and S-adenosyl-L-homocysteine | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Thompson, A.A, Geiss, B.J, Peersen, O.B. | | Deposit date: | 2008-10-13 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Analysis of flavivirus NS5 methyltransferase cap binding.
J.Mol.Biol., 385, 2009
|
|
4TN2
 
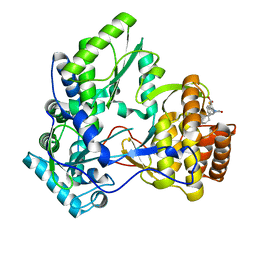 | | NS5b in complex with lactam-thiophene carboxylic acids | | Descriptor: | 3-[(2R)-2-cyclohexyl-5-oxopyrrolidin-1-yl]-5-phenylthiophene-2-carboxylic acid, Genome polyprotein | | Authors: | Chopra, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of lactam-thiophene carboxylic acids as potent hepatitis C virus polymerase inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
3EVC
 
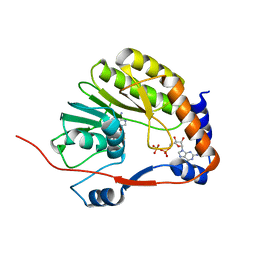 | |
8VLB
 
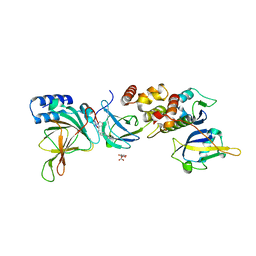 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 4 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
8VL9
 
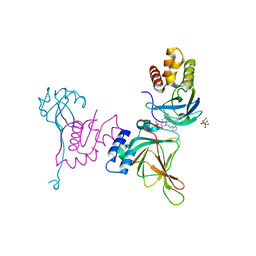 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 8 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
7V1F
 
 | |
1B72
 
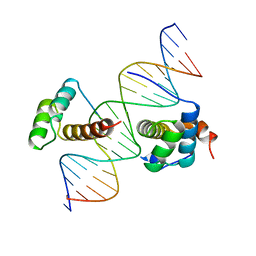 | | PBX1, HOMEOBOX PROTEIN HOX-B1/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*GP*AP*TP*CP*GP*GP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*CP*GP*AP*TP*CP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3'), PROTEIN (HOMEOBOX PROTEIN HOX-B1), ... | | Authors: | Piper, D.E, Batchelor, A.H, Chang, C.-P, Cleary, M.L, Wolberger, C. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation.
Cell(Cambridge,Mass.), 96, 1999
|
|
5X7X
 
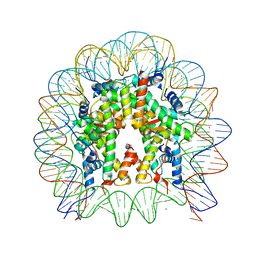 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
5G5P
 
 | | Structure of the Saccharomyces cerevisiae TREX-2 complex | | Descriptor: | 26S PROTEASOME COMPLEX SUBUNIT SEM1, NUCLEAR MRNA EXPORT PROTEIN SAC3, NUCLEAR MRNA EXPORT PROTEIN THP1 | | Authors: | Aibara, S, Bai, X.C, Stewart, M. | | Deposit date: | 2016-05-26 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The Sac3 Tpr-Like Region in the Saccharomyces Cerevisiae Trex-2 Complex is More Extensive But Independent of the Cid Region
J.Struct.Biol., 195, 2016
|
|
3EVD
 
 | |
2W9B
 
 | | Binary complex of Dpo4 bound to N2,N2-dimethyl-deoxyguanosine modified DNA | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOCP)-3', 5'-D(*TP*CP*AP*TP*M2GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', DNA POLYMERASE IV, ... | | Authors: | Eoff, R.L, Zhang, H, Kosekov, I.D, Rizzo, C.J, Egli, M, Guengerich, F.P. | | Deposit date: | 2009-01-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Function Relationships in Miscoding by Sulfolobus Solfataricus DNA Polymerase Dpo4: Guanine N2,N2-Dimethyl Substitution Produces Inactive and Miscoding Polymerase Complexes.
J.Biol.Chem., 284, 2009
|
|
5R8Y
 
 | |
5R9F
 
 | |
5R92
 
 | |
5R9I
 
 | |
5R9W
 
 | |
