3TF5
 
 | |
1BCX
 
 | |
1NSK
 
 | |
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
1TE7
 
 | | Solution NMR Structure of Protein yqfB from Escherichia coli. Northeast Structural Genomics Consortium Target ET99 | | Descriptor: | Hypothetical UPF0267 protein yqfB | | Authors: | Atreya, H.S, Shen, Y, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | G-Matrix Fourier Transform NOESY-Based Protocol for High-Quality Protein Structure Determination
J.Am.Chem.Soc., 127, 2005
|
|
3OC3
 
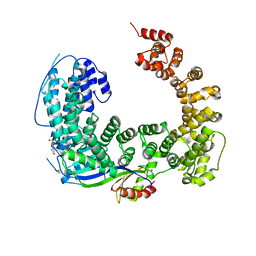 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
6SMU
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4I3W
 
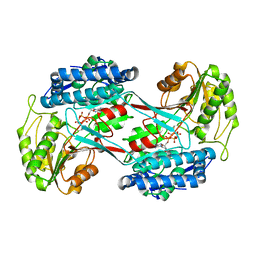 | |
4HXQ
 
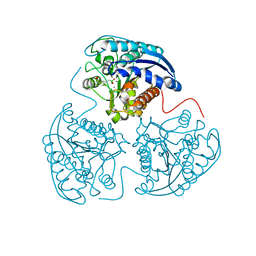 | | Crystal structure of human Arginase-1 complexed with inhibitor 14 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-carboxy-5-(methylamino)-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
1T02
 
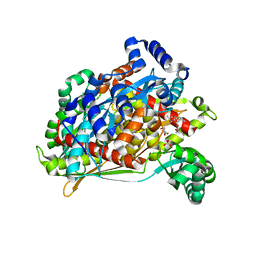 | | Crystal structure of a Statin bound to class II HMG-CoA reductase | | Descriptor: | (3R,5R)-7-((1R,2R,6S,8R,8AS)-2,6-DIMETHYL-8-{[(2R)-2-METHYLBUTANOYL]OXY}-1,2,6,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL)-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Tabernero, L, Rodwell, V.W, Stauffacher, C. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a statin bound to a class II hydroxymethylglutaryl-CoA reductase.
J.Biol.Chem., 278, 2003
|
|
3SAW
 
 | | MUTM Slanted complex 8 with R112A mutation | | Descriptor: | 5'-D(*A*GP*GP*TP*AP*GP*AP*CP*AP*AP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*C*CP*TP*TP*GP*TP*(CX2)P*TP*AP*CP*C)-3', DNA GLYCOSYLASE, ... | | Authors: | Qi, Y, Verdine, G.L. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8XPM
 
 | | Mature virion portal of phage lambda with DNA | | Descriptor: | DNA (104-MER), DNA (92-MER), Head completion protein, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
3US2
 
 | |
8XOT
 
 | | Prohead portal of bacteriophage lambda | | Descriptor: | Portal protein B | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
4I1O
 
 | | Crystal structure of the Legionella pneumophila GAP domain of LepB in complex with Rab1b bound to GDP and BeF3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gazdag, E.M, Streller, A, Vetter, I.R, Goody, R.S, Itzen, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB.
Embo Rep., 14, 2013
|
|
3OD1
 
 | | The crystal structure of an ATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C | | Descriptor: | ATP phosphoribosyltransferase regulatory subunit, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Bigelow, L, Hamilton, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of aATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C
To be Published
|
|
3SCS
 
 | | Crystal Structure of Rice BGlu1 E386S Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
4GO4
 
 | | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative gamma-hydroxymuconic semialdehyde dehydrogenase | | Authors: | Su, J, Zhang, C, Liu, S, Zhu, D, Gu, L. | | Deposit date: | 2012-08-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide
To be Published
|
|
1T0T
 
 | | Crystallographic structure of a putative chlorite dismutase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, APC35880, MAGNESIUM ION | | Authors: | Gilski, M, Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of APC35880 protein from Bacillus Stearothermophilus
To be Published
|
|
4GRV
 
 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
3NKN
 
 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
1T47
 
 | | Structure of fe2-HPPD bound to NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Brownlee, J, Johnson-Winters, K, Harrison, D.H.T, Moran, G.R. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ferrous Form of (4-Hydroxyphenyl)pyruvate Dioxygenase from Streptomyces avermitilis in Complex with the Therapeutic Herbicide, NTBC
Biochemistry, 43, 2004
|
|
1T4T
 
 | | arginase-dinor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}ALANINE, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1T2E
 
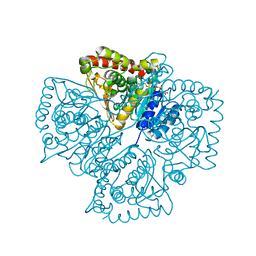 | | Plasmodium falciparum lactate dehydrogenase S245A, A327P mutant complexed with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
3N8W
 
 | |
