6CJR
 
 | | Candida albicans Hsp90 nucleotide binding domain in complex with SNX-2112 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein 90 homolog | | Authors: | Kirkpatrick, M.G, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
5N5K
 
 | |
6F1Z
 
 | | Roadblock-1 region of the dynein tail/dynactin/BICDR1 complex | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Dynein light chain roadblock-type 1 | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
8BWB
 
 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
6FF4
 
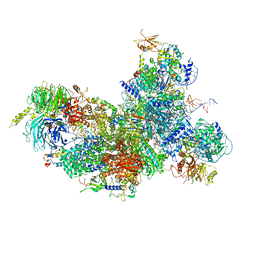 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
1YGJ
 
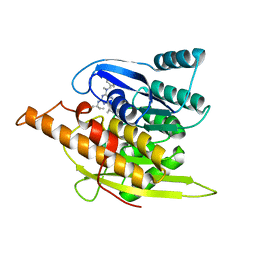 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | (2R)-2-({6-[BENZYL(METHYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)BUTAN-1-OL, Pyridoxal kinase | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
6B3E
 
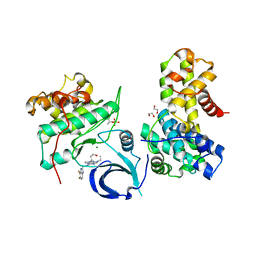 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
1MAA
 
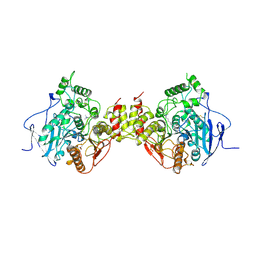 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
6AZE
 
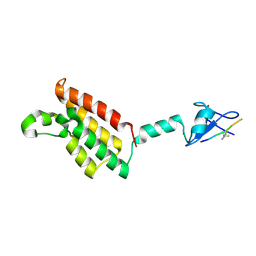 | |
5N86
 
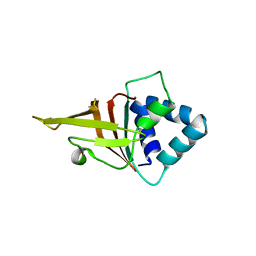 | | Crystal structure of FAS1 domain of hyaluronic acid receptor stabilin-2 | | Descriptor: | Stabilin-2 | | Authors: | Twarda-Clapa, A, Labuzek, B, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Crystal structure of the FAS1 domain of the hyaluronic acid receptor stabilin-2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4GUX
 
 | | Crystal structure of trypsin:MCoTi-II complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cationic trypsin, ... | | Authors: | King, G.J, Daly, N.L, Thorstholm, L, Greenwood, K.P, Rosengren, K.J, Heras, B, Craik, D.J, Martin, J.L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor
J.Biol.Chem., 288, 2013
|
|
1YGK
 
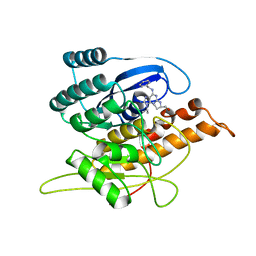 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | Pyridoxal kinase, R-ROSCOVITINE | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
5IAS
 
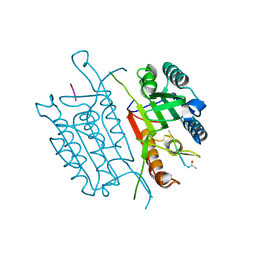 | | Caspase 3 V266Y | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6CJS
 
 | |
1YHJ
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | (2R)-2-{[6-(BENZYLOXY)-9-ISOPROPYL-9H-PURIN-2-YL]AMINO}BUTAN-1-OL, Pyridoxal Kinase | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-09 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives.
J.Biol.Chem., 280, 2005
|
|
1ZGA
 
 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-6a-hydroxymaackiain | | Descriptor: | (6AR,12AR)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMENE-3,6A(12AH)-DIOL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
6DIK
 
 | | Crystal structure of Bothropstoxin I (BthTX-I) complexed to Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, BICARBONATE ION, Basic phospholipase A2 homolog bothropstoxin-1, ... | | Authors: | Cardoso, F.F, Salvador, G.H.M, Borges, R.J. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis of phospholipase A2-like myotoxin inhibition by chicoric acid, a novel potent inhibitor of ophidian toxins.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
4FZ1
 
 | |
4ZYI
 
 | |
5I9B
 
 | | Caspase 3 V266A | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8DIW
 
 | | Crystal structure of NavAb E96P as a basis for the human Nav1.7 Inherited Erythromelalgia S211P mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
5IBC
 
 | | Caspase 3 V266I | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8DIV
 
 | | Crystal structure of NavAb I22V as a basis for the human Nav1.7 Inherited Erythromelalgia I136V mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein, ... | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Powell, N.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIX
 
 | | Structure of NavAb L98R as a basis for the human Nav1.7 Inherited Erythromelalgia L823R mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
