5LK6
 
 | |
5LRF
 
 | | Crystal structure of Glycogen Phosphorylase b in complex with KS389 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-06-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | van der Waals interactions govern C-beta-d-glucopyranosyl triazoles' nM inhibitory potency in human liver glycogen phosphorylase.
J. Struct. Biol., 199, 2017
|
|
7PQR
 
 | | LsAA9A expressed in E. coli | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Muderspach, S.J, Metherall, J, Ipsen, J, Rollan, C.H, Norholm, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7R3X
 
 | | The crystal structure of the L439V variant of Pol2CORE in complex with DNA and an incoming nucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ACETATE ION, CALCIUM ION, ... | | Authors: | Barbari, S.R, Beach, A.K, Markgren, J.G, Parkash, V, Johansson, E, Shcherbakova, P.V. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ε variants at low dNTP levels.
Nucleic Acids Res., 50, 2022
|
|
5LRE
 
 | | Crystal structure of Glycogen Phosphorylase b in complex with KS382 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-31 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur J Med Chem, 123, 2016
|
|
5LRD
 
 | | Crystal structure of Glycogen Phosphorylase b in complex with KS242 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-methylphenyl)-4~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-06-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | van der Waals interactions govern C-beta-d-glucopyranosyl triazoles' nM inhibitory potency in human liver glycogen phosphorylase.
J. Struct. Biol., 199, 2017
|
|
8Z62
 
 | |
3K1S
 
 | | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PTS system, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis
To be Published
|
|
4LHP
 
 | | Crystal Structure of Native FG41Malonate Semialdehyde Decarboxylase | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
8S3E
 
 | | Structure of rabbit Slo1 in complex with gamma1/LRRC26 | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Redhardt, M, Raunser, S, Raisch, T. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Cryo-EM structure of the Slo1 potassium channel with the auxiliary gamma 1 subunit suggests a mechanism for depolarization-independent activation.
Febs Lett., 598, 2024
|
|
4LES
 
 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3K3J
 
 | | P38alpha bound to novel DFG-out compound PF-00416121 | | Descriptor: | 2-(4-fluorophenyl)-3-oxo-6-pyridin-4-yl-N-[2-(trifluoromethyl)benzyl]-2,3-dihydropyridazine-4-carboxamide, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | The Design, Synthesis and Potential Utility of Fluorescence Probes that Target DFG-out Conformation of p38alpha for High Throughput Screening Binding Assay.
Chem.Biol.Drug Des., 74, 2009
|
|
3D7Z
 
 | |
8QEF
 
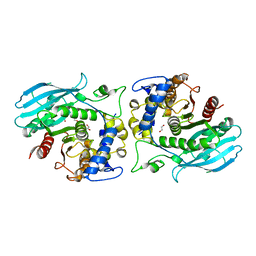 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola ATCC 8482 bound to novel ligand. | | Descriptor: | 1,2-ETHANEDIOL, Putative acetyl xylan esterase, beta-D-galactopyranuronic acid | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
8P1E
 
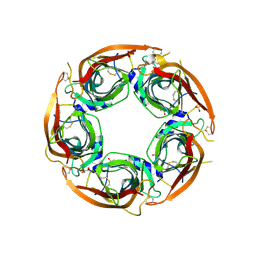 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001613. | | Descriptor: | 1-[4-(trifluoromethyl)pyridin-2-yl]piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8P22
 
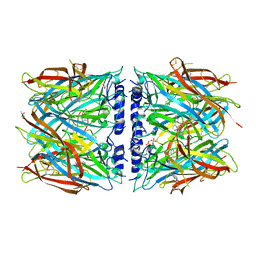 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with IOTA376. | | Descriptor: | 2-[(2~{R})-1-ethylimidazolidin-2-yl]-6-pyridin-2-yl-pyridine, Acetylcholine-binding protein, GLYCEROL, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-14 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8VXE
 
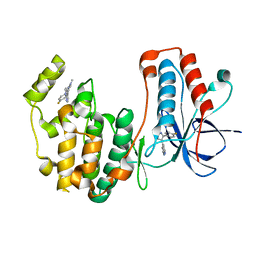 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8QKA
 
 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK9
 
 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OSL
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8R2J
 
 | |
8R38
 
 | | BIIG2 anti-integrin Fab | | Descriptor: | BIIG2 Fab, heavy chain, light chain, ... | | Authors: | Cordara, G, Heim, J.B, Johannesen, H, Krengel, U. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-25 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural and functional characterization of integrin alpha 5-targeting antibodies for anti-angiogenic therapy.
Biorxiv, 2025
|
|
6COI
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I with CYANIDE, benzaldehyde, MANDELIC ACID NITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, (S)-MANDELIC ACID NITRILE, Alpha-hydroxynitrile lyase, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.024 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
