6M6T
 
 | | Amylomaltase from Streptococcus agalactiae in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Wangkanont, K, Tumhom, S, Pongsawasdi, P. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Streptococcus agalactiae amylomaltase offers insight into the transglycosylation mechanism and the molecular basis of thermostability among amylomaltases.
Sci Rep, 11, 2021
|
|
8E4T
 
 | | Crystal structure of the kinase domain of RTKC8 from the choanoflagellate Monosiga brevicollis | | Descriptor: | PHOSPHATE ION, RTKC8 Kinase domain, STAUROSPORINE | | Authors: | Bajaj, T, Gee, C.L, Kuriyan, J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the kinase domain of a receptor tyrosine kinase from a choanoflagellate, Monosiga brevicollis.
Plos One, 18, 2023
|
|
6US6
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6PR4
 
 | | D262N/S128A S. typhimurium siroheme synthase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
8E8X
 
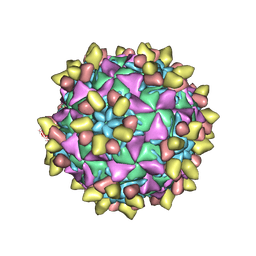 | | 9H2 Fab-Sabin poliovirus 3 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
6PRO
 
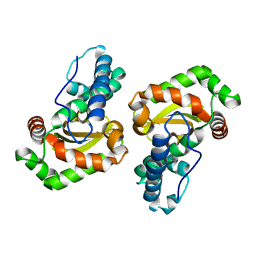 | | MnSOD from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Adams, J.J, Morton, C.J, Parker, M.W. | | Deposit date: | 2019-07-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | The Crystal Structure of the Manganese Superoxide Dismutase from Geobacillus stearothermophilus: Parker and Blake (1988) Revisited
Aust.J.Chem., 73, 2020
|
|
6M2D
 
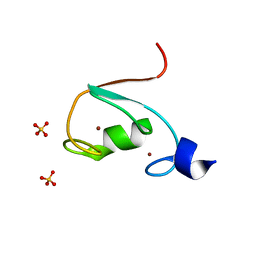 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
7MRJ
 
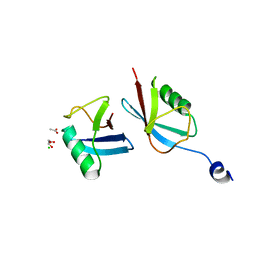 | | Crystal structure of a novel ubiquitin-like TINCR | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Forouhar, F, Morgado-Palacin, L, Brown, J.A, Martinez, T, Pedrero, J.M.G, Reglero, C, Chaudhry, I, Vaughan, J, Rodriguez-Perales, S, Allonca, E, Granda-Diaz, R, Quinn, S.A, Fernandez, A.F, Fraga, M.F, Kim, A.L, Santos-Juanes, J, Owens, D.M, Rodrigo, J.P, Saghatelian, A, Ferrando, A.A. | | Deposit date: | 2021-05-07 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The TINCR ubiquitin-like microprotein is a tumor suppressor in squamous cell carcinoma.
Nat Commun, 14, 2023
|
|
6UVI
 
 | |
6M2C
 
 | |
6PRZ
 
 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6UVS
 
 | |
7M0C
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound mismatched DSB substrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
8DNG
 
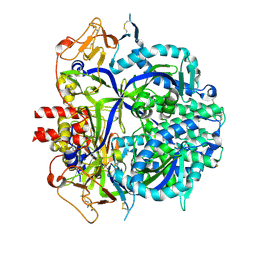 | |
6PHH
 
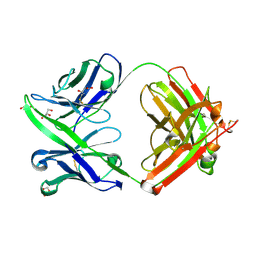 | |
6LRM
 
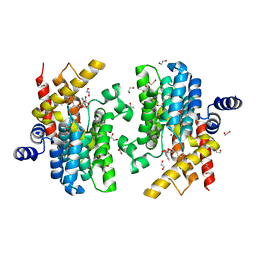 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
8DMG
 
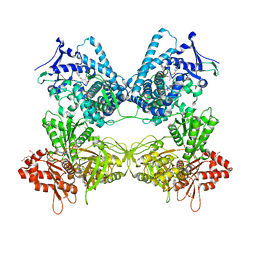 | | CYP102A1 in Closed Conformation | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional cytochrome P450/NADPH--P450 reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Su, M, Xu, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Insight into the conformational dynamics of cytochrome P450 CYP102A1 enzyme using Cryo-EM
To Be Published
|
|
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MQZ
 
 | | Cytochrome c oxidase assembly factor 7 | | Descriptor: | Cytochrome c oxidase assembly factor 7 | | Authors: | Maghool, S, Maher, M.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Mitochondrial COA7 is a heme-binding protein with disulfide reductase activity, which acts in the early stages of complex IV assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6M6M
 
 | | The crystal structure of glycosidase mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase | | Authors: | Wang, R.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization and Structural Elucidation of Enhanced Catalytic Activity upon Engineering Glycosidase KfGH01 for the Production of Vina-ginsenoside R7
To Be Published
|
|
6OYU
 
 | | Structure of an ancestral-reconstructed cytochrome P450 1B1 with alpha-naphthoflavone | | Descriptor: | 2-PHENYL-4H-BENZO[H]CHROMEN-4-ONE, Cytochrome P450 1B1, GLYCEROL, ... | | Authors: | Bart, A.G, Harris, K.L, Scott, E.E. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of an ancestral mammalian family 1B1 cytochrome P450 with increased thermostability.
J.Biol.Chem., 295, 2020
|
|
6USF
 
 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
7M3V
 
 | | RNA bulged-G motif | | Descriptor: | IRIDIUM (III) ION, RNA (27-MER) | | Authors: | Kondo, J, Sekiguchi, S. | | Deposit date: | 2021-03-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | RNA bulged-G motif
To Be Published
|
|
