2IDO
 
 | | Structure of the E. coli Pol III epsilon-Hot proofreading complex | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III epsilon subunit, Hot protein, ... | | Authors: | Kirby, T.W, Harvey, S, DeRose, E.F, Chalov, S, Chikova, A.K, Perrino, F.W, Schaaper, R.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex.
J.Biol.Chem., 281, 2006
|
|
1U59
 
 | | Crystal Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ZAP-70 | | Authors: | Jin, L, Pluskey, S, Petrella, E.C, Cantin, S.M, Gorga, J.C, Rynkiewicz, M.J, Pandey, P, Strickler, J.E, Babine, R.E, Weaver, D.T, Seidl, K.J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine: IMPLICATIONS FOR THE DESIGN OF SELECTIVE INHIBITORS
J.Biol.Chem., 279, 2004
|
|
4D1E
 
 | | THE CRYSTAL STRUCTURE OF HUMAN MUSCLE ALPHA-ACTININ-2 | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Pinotsis, N, Salmazo, A, Sjoeblom, B, Gkougkoulia, E, Djinovic-Carugo, K. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure and Regulation of Human Muscle Alpha-Actinin
Cell(Cambridge,Mass.), 159, 2014
|
|
5D5M
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2015-08-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
3SH0
 
 | |
6Z3T
 
 | | Structure of canine Sec61 inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Gerard, S.F, Higgins, M.K. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Inhibited State of the Sec Translocon.
Mol.Cell, 79, 2020
|
|
4WA9
 
 | |
2ITM
 
 | | Crystal structure of the E. coli xylulose kinase complexed with xylulose | | Descriptor: | AMMONIUM ION, D-XYLULOSE, SULFATE ION, ... | | Authors: | di Luccio, E, Voegtli, J, Wilson, D.K. | | Deposit date: | 2006-10-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli.
J.Mol.Biol., 365, 2007
|
|
2IV2
 
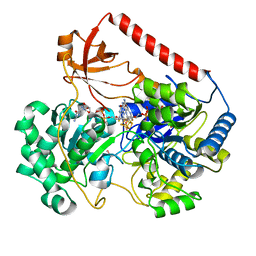 | | Reinterpretation of reduced form of formate dehydrogenase H from E. coli | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase H, GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, ... | | Authors: | Raaijmakers, H.C.A, Romao, M.J. | | Deposit date: | 2006-06-08 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Formate-Reduced E. Coli Formate Dehydrogenase H: The Reinterpretation of the Crystal Structure Suggests a New Reaction Mechanism.
J.Biol.Inorg.Chem., 11, 2006
|
|
6EXN
 
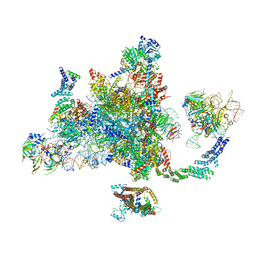 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
1U94
 
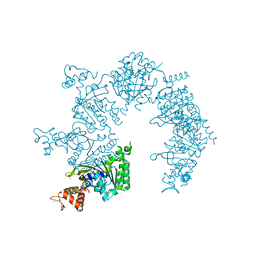 | |
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8HI1
 
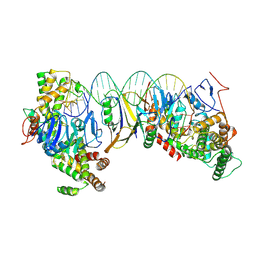 | | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, DNA (26-MER), DNA (31-MER), ... | | Authors: | Chen, Q, Luo, Y. | | Deposit date: | 2022-11-18 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism.
Innovation (N Y), 4, 2023
|
|
4CKK
 
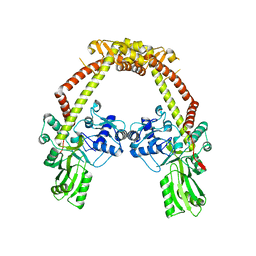 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
2HZA
 
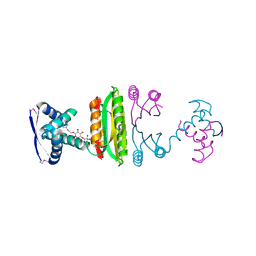 | | Nickel-bound full-length Escherichia coli NikR | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulator | | Authors: | Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NikR-operator complex structure and the mechanism of repressor activation by metal ions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6YF2
 
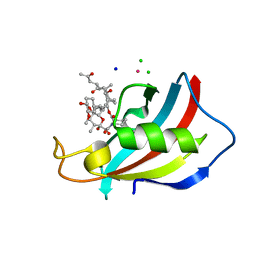 | | FKBP12 in complex with the BMP potentiator compound 6 at 1.03A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-23,25-dimethoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14-bis(oxidanyl)-17-(2-oxidanylidenepropyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF3
 
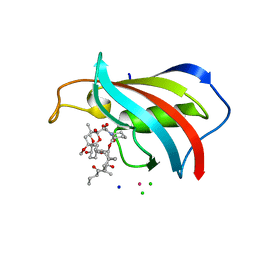 | | FKBP12 in complex with the BMP potentiator compound 10 at 1.00A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-17-ethyl-25-methoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14,23-tris(oxidanyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
2VHJ
 
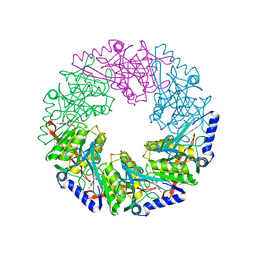 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
4UZL
 
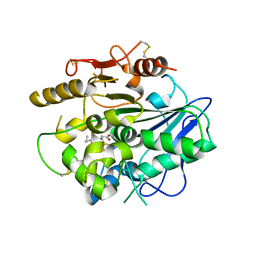 | |
2I6R
 
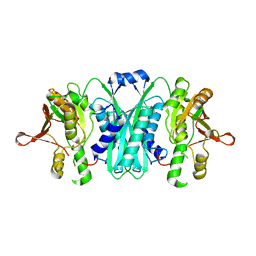 | | Crystal structure of E. coli HypE, a hydrogenase maturation protein | | Descriptor: | HypE protein | | Authors: | Rangarajan, E.S, Proteau, A, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
3S2Y
 
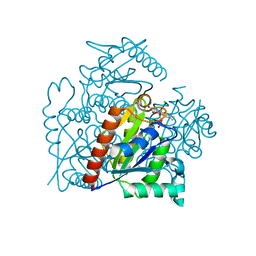 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
2VHT
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 R279A mutant in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
3CX2
 
 | | Crystal structure of the C1 domain of cardiac isoform of myosin binding protein-C at 1.3A | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Fisher, S.J, Helliwell, J.R, Khurshid, S, Govada, L, Redwood, C, Squire, J.M, Chayen, N.E. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1TOR
 
 | | MOLECULAR DYNAMICS SIMULATION FROM 2D-NMR DATA OF THE FREE ACHR MIR DECAPEPTIDE AND THE ANTIBODY-BOUND [A76]MIR ANALOGUE | | Descriptor: | ACETYLCHOLINE RECEPTOR, MAIN IMMUNOGENIC REGION | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
8ACW
 
 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.4 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
