4KAN
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)acetic acid, ADENOSINE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid
To be Published
|
|
4KD4
 
 | | Crystal structure of Staphylococcal nuclease variant V23L/L25V/V66I/I72V at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
3K0A
 
 | | Crystal structure of the phosphorylation-site mutant S431A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3JAG
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
4KE7
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-myristoyl glycerol analogue | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase, dodecyl hydrogen (S)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KFM
 
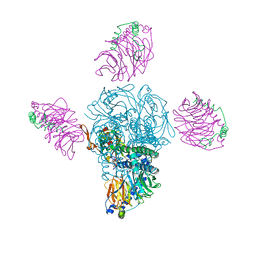 | |
3JBV
 
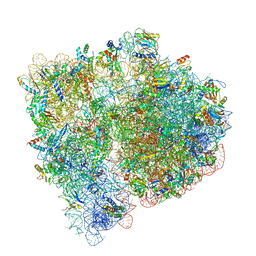 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
4KE1
 
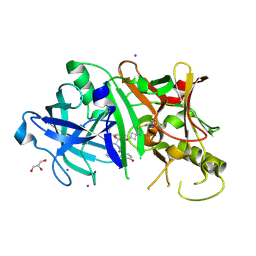 | | Crystal structure of BACE1 in complex with hydroxyethylamine-macrocyclic inhibitor 19 | | Descriptor: | (12S)-12-[(1R)-2-{[(4S)-6-ethyl-3,4-dihydrospiro[chromene-2,1'-cyclobutan]-4-yl]amino}-1-hydroxyethyl]-1,13-diazatricyclo[13.3.1.1~6,10~]icosa-6(20),7,9,15(19),16-pentaene-14,18-dione, Beta-Secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M, Li, V. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KGV
 
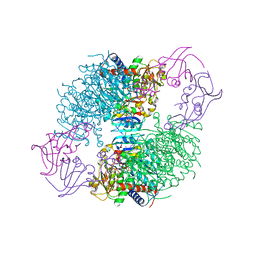 | | The R state structure of E. coli ATCase with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
4KHM
 
 | | HCV NS5B GT1A with GSK5852 | | Descriptor: | HCV Polymerase, SULFATE ION, [4-({[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}methyl)-2-fluorophenyl]boronic acid | | Authors: | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
4KEA
 
 | | Crystal structure of D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in space group P212121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3JSI
 
 | | Human phosphodiesterase 9 in complex with inhibitor | | Descriptor: | 6-benzyl-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2009-09-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of a Brain Penetrant PDE9A Inhibitor Utilizing Prospective Design and Chemical Enablement as a Rapid Lead Optimization Strategy.
J.Med.Chem., 52, 2009
|
|
4KK2
 
 | | Crystal structure of a chimeric FPP/GFPP synthase (TARGET EFI-502313c) from Artemisia spiciformiS (1-72:GI751454468,73-346:GI75233326), apo structure | | Descriptor: | Monoterpene synthase FDS-5, chloroplastic - Farnesyl diphosphate synthase 1 chimera | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-05-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a chimeric FPP/GFPP synthase (TARGET EFI-502313c) from Artemisia spiciformis (1-72:GI751454468,73-346:GI75233326), apo structure
To be Published
|
|
4KLM
 
 | | DNA polymerase beta matched product complex with Mg2+, 11 h | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
3J4Q
 
 | |
4KMO
 
 | | Crystal Structure of the Vps33-Vps16 HOPS subcomplex from Chaetomium thermophilum | | Descriptor: | Putative vacuolar protein sorting-associated protein, SULFATE ION, Small conjugating protein ligase-like protein | | Authors: | Baker, R.W, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Plos One, 8, 2013
|
|
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KNM
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with 2-Chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 13, TRIETHYLENE GLYCOL, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
4KKX
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium with 2-aminophenol quinonoid in the beta site and the F6 inhibitor in the alpha site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2013-05-06 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
4KP3
 
 | | Crystal Structure of MyoVa-GTD in Complex with Two Cargos | | Descriptor: | Melanophilin, RILP-like protein 2, Unconventional myosin-Va | | Authors: | Wei, Z, Liu, X, Yu, C, Zhang, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J70
 
 | | Model of gp120, including variable regions, in complex with CD4 and 17b | | Descriptor: | Envelope glycoprotein gp120, T-cell surface glycoprotein CD4, envelope glycoprotein gp41, ... | | Authors: | Rasheed, M, Bettadapura, R, Bajaj, C. | | Deposit date: | 2014-04-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Computational Refinement and Validation Protocol for Proteins with Large Variable Regions Applied to Model HIV Env Spike in CD4 and 17b Bound State.
Structure, 23, 2015
|
|
4KRA
 
 | |
4K39
 
 | | Native anSMEcpe with bound AdoMet and Cp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, Cp18Cys peptide, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.275 Å) | | Cite: | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
4K3L
 
 | | E. coli sliding clamp in complex with AcLF dipeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
