4YS6
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE | | Descriptor: | CHLORIDE ION, Putative solute-binding component of ABC transporter, SODIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE
To be published
|
|
4MUR
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUQ
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.364 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4YWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE | | Descriptor: | ABC TRANSPORTER SOLUTE BINDING PROTEIN, beta-D-xylopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE
To be published
|
|
4MUS
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4YI5
 
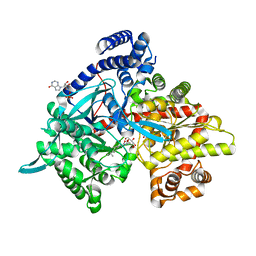 | | Crystal structure of Gpb in complex with 4b | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Stravodimos, G.A, Leonidas, D.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycogen phosphorylase as a target for type 2 diabetes: synthetic, biochemical, structural and computational evaluation of novel N-acyl-N -( beta-D-glucopyranosyl) urea inhibitors.
Curr Top Med Chem, 15, 2015
|
|
227D
 
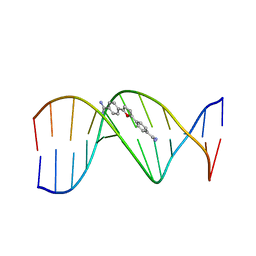 | | A CRYSTALLOGRAPHIC AND SPECTROSCOPIC STUDY OF THE COMPLEX BETWEEN D(CGCGAATTCGCG)2 AND 2,5-BIS(4-GUANYLPHENYL)FURAN, AN ANALOGUE OF BERENIL. STRUCTURAL ORIGINS OF ENHANCED DNA-BINDING AFFINITY | | Descriptor: | 2,5-BIS(4-GUANYLPHENYL)FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Laughton, C.A, Tanious, F, Nunn, C.M, Boykin, D.W, Wilson, W.D, Neidle, S. | | Deposit date: | 1995-08-08 | | Release date: | 1995-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity.
Biochemistry, 35, 1996
|
|
9XIA
 
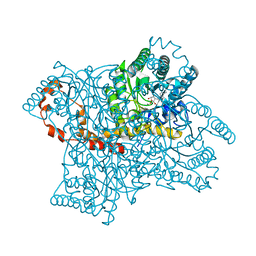 | |
4RW5
 
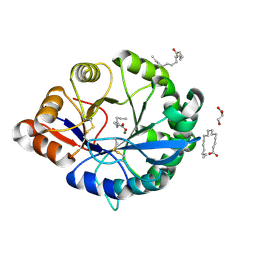 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-TRIDECANOIC ACID, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
4TWC
 
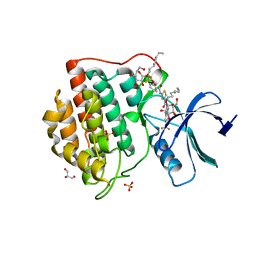 | | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4- carboxamide derivatives as potent inhibitors of CK1d/e | | Descriptor: | 2-{[2-(trifluoromethoxy)benzoyl]amino}-N-[6-(trifluoromethyl)-1H-benzimidazol-2-yl]-1,3-thiazole-4-carboxamide, Casein kinase I isoform delta, DIMETHYL SULFOXIDE, ... | | Authors: | Bischof, J, Leban, L, Zaja, M, Grothey, A, Radunsky, B, Othersen, O, Strobl, S, Vitt, D, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1d/e.
Amino Acids, 43, 2012
|
|
4U8U
 
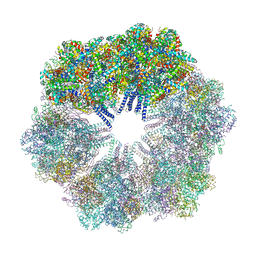 | | The Crystallographic structure of the giant hemoglobin from Glossoscolex paulistus at 3.2 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Bachega, J.F.R, Maluf, F.V, Andi, B, D'Muniz Pereira, H, Carazzollea, M.F, Orville, A, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-08-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UAG
 
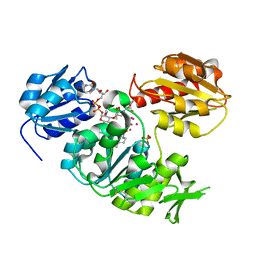 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
4X56
 
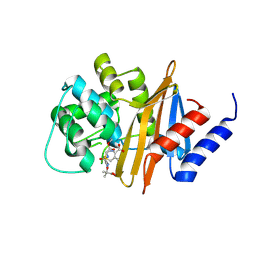 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4X53
 
 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BICARBONATE ION, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
7RN3
 
 | | hyen D solution structure | | Descriptor: | Cyclotide hyen-D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Mutagenesis of bracelet cyclotide hyen D reveals functionally and structurally critical residues for membrane binding and cytotoxicity.
J.Biol.Chem., 298, 2022
|
|
8BZI
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
4XTU
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide) | | Descriptor: | Bifunctional ligase/repressor BirA, N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6500299 Å) | | Cite: | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide)
To be Published
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6GVA
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR4455 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-azanylethylsulfanyl)-3-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, BROMIDE ION, ... | | Authors: | Skerlova, J, Rezacova, P. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d]pyrimidine Inhibitors of Cyclin-Dependent Kinases and Their Evaluation in Lymphoma Models.
J.Med.Chem., 62, 2019
|
|
7S04
 
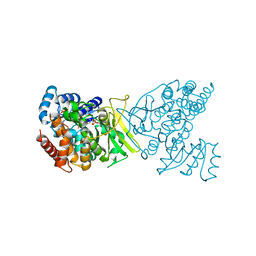 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (IspC) from Acinetobacter baumannii in complex with FR900098, NADPH, and a magnesium ion | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, MAGNESIUM ION, ... | | Authors: | Noble, S.M, Ball, H.S, Couch, R.D. | | Deposit date: | 2021-08-28 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Characterization and Inhibition of 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase: A Promising Drug Target in Acinetobacter baumannii and Klebsiella pneumoniae .
Acs Infect Dis., 7, 2021
|
|
4WUT
 
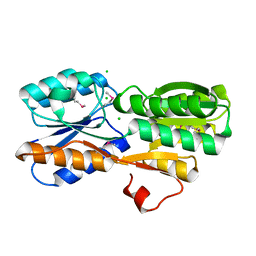 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
8BSA
 
 | | Vc1313-LBD bound to D-arginine | | Descriptor: | D-ARGININE, Methyl-accepting chemotaxis protein | | Authors: | ter Beek, J, Berntsson, R.P.-A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-amino acids signal a stress-dependent run-away response in Vibrio cholerae.
Nat Microbiol, 8, 2023
|
|
4WWH
 
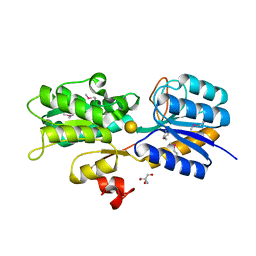 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, beta-D-galactopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE
To be published
|
|
4XFE
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, SULFATE ION, TRAP dicarboxylate transporter subunit DctP, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate
To be published
|
|
8BSB
 
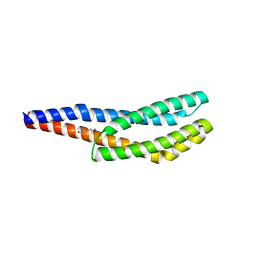 | | Vc1313-LBD bound to D-lysine | | Descriptor: | D-LYSINE, Methyl-accepting chemotaxis protein | | Authors: | ter Beek, J, Berntsson, R.P.-A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-amino acids signal a stress-dependent run-away response in Vibrio cholerae.
Nat Microbiol, 8, 2023
|
|
