4GBZ
 
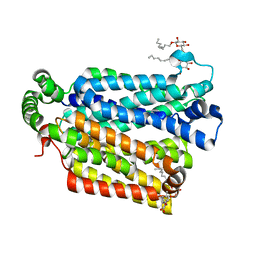 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | Descriptor: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GC0
 
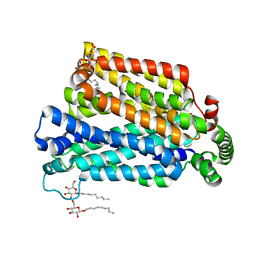 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
8Y66
 
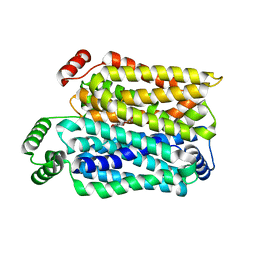 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y65
 
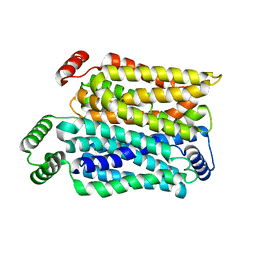 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8BVS
 
 | | Cryo-EM structure of rat SLC22A6 bound to tenofovir | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody), ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVR
 
 | | Cryo-EM structure of rat SLC22A6 in the apo state | | Descriptor: | PHOSPHATE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody) | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVT
 
 | | Cryo-EM structure of rat SLC22A6 bound to probenecid | | Descriptor: | 4-(dipropylsulfamoyl)benzoic acid, Solute carrier family 22 member 6, Synthetic nanobody (Sybody) | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BW7
 
 | | Cryo-EM structure of rat SLC22A6 bound to alpha-ketoglutaric acid | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Solute carrier family 22 member 6, ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ET7
 
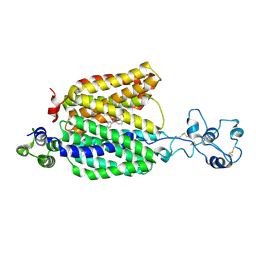 | | Cryo-EM structure of the organic cation transporter 1 in complex with diphenhydramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, OCT1 | | Authors: | Suo, Y, Wright, N.J, Lee, S.-Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Molecular basis of polyspecific drug and xenobiotic recognition by OCT1 and OCT2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ET8
 
 | | Cryo-EM structure of the organic cation transporter 1 in complex with verapamil | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OCT1 | | Authors: | Suo, Y, Wright, N.J, Lee, S.-Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Molecular basis of polyspecific drug and xenobiotic recognition by OCT1 and OCT2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ET9
 
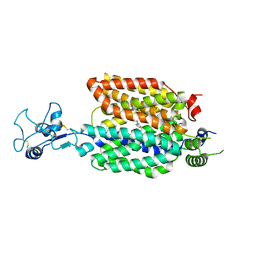 | |
8ET6
 
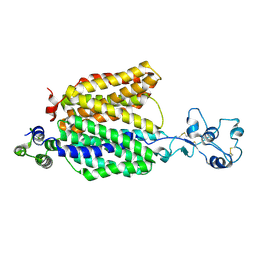 | |
8JTS
 
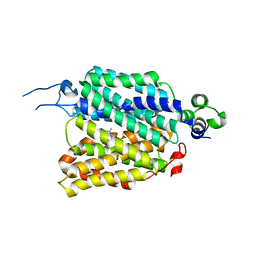 | | hOCT1 in complex with metformin in outward open conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTW
 
 | | hOCT1 in complex with nb5660 in inward facing partially open 1 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTX
 
 | | hOCT1 in complex with nb5660 in inward facing fully open conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTT
 
 | | hOCT1 in complex with metformin in outward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTV
 
 | | hOCT1 in complex with metformin in inward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
4YBQ
 
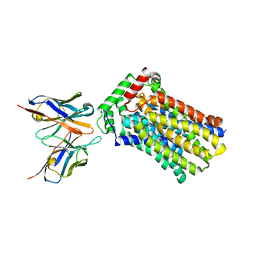 | | Rat GLUT5 with Fv in the outward-open form | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5, antibody Fv fragment heavy chain, ... | | Authors: | Nomura, N, Shimamura, T, Iwata, S. | | Deposit date: | 2015-02-19 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5
Nature, 526, 2015
|
|
4YB9
 
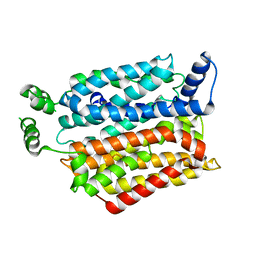 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
4ZWB
 
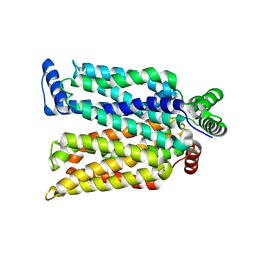 | | Crystal structure of maltose-bound human GLUT3 in the outward-occluded conformation at 2.4 angstrom | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 3, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZWC
 
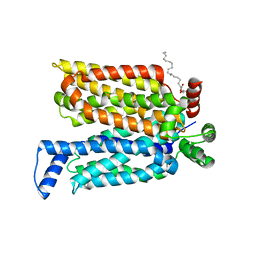 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZW9
 
 | | Crystal structure of human GLUT3 bound to D-glucose in the outward-occluded conformation at 1.5 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
8SC3
 
 | |
8SC4
 
 | |
8SDU
 
 | | Structure of rat organic anion transporter 1 (OAT1) | | Descriptor: | Solute carrier family 22 member 6 | | Authors: | Dou, T, Jiang, J. | | Deposit date: | 2023-04-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | The substrate and inhibitor binding mechanism of polyspecific transporter OAT1 revealed by high-resolution cryo-EM.
Nat.Struct.Mol.Biol., 30, 2023
|
|
