7Z0O
 
 | | Structure of transcription factor UAF in complex with TBP and 35S rRNA promoter DNA | | Descriptor: | Histone H3, Histone H4, Non-template DNA, ... | | Authors: | Baudin, F, Murciano, B, Fung, H.K.H, Fromm, S.A, Mueller, C.W. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of RNA polymerase I selection by transcription factor UAF.
Sci Adv, 8, 2022
|
|
3Q33
 
 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
6J2P
 
 | |
2YGV
 
 | | Conserved N-terminal domain of the yeast Histone Chaperone Asf1 in complex with the C-terminal fragment of Rad53 | | Descriptor: | GLYCEROL, HISTONE CHAPERONE ASF1, SERINE/THREONINE-PROTEIN KINASE RAD53, ... | | Authors: | Jiao, Y, Seeger, K, Murciano, B, Ledu, M.H, Charbonnier, J.B, Legrand, P, Lautrette, A, Gaubert, A, Mousson, F, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Surprising Complexity of the Asf1 Histone Chaperone-Rad53 Kinase Interaction
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1WG3
 
 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
7UQJ
 
 | | Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ATPgS and histone H3 tail in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase histone chaperone YTA7, Histone H3, ... | | Authors: | Wang, F, Feng, X, Li, H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly.
J.Biol.Chem., 299, 2022
|
|
4JJN
 
 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7XAY
 
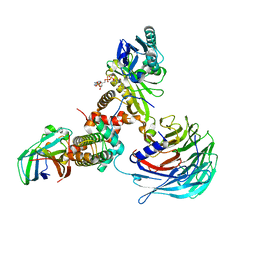 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
7UNK
 
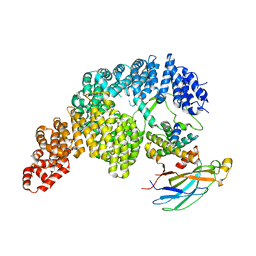 | | Structure of Importin-4 bound to the H3-H4-ASF1 histone-histone chaperone complex | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Bernardes, N.E, Chook, Y.M, Fung, H.Y.J, Chen, Z, Li, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E9C
 
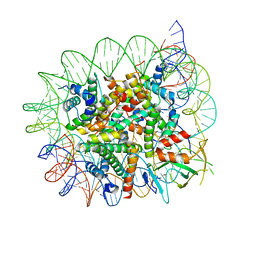 | | Cryo-EM structure of the 1:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
6GEJ
 
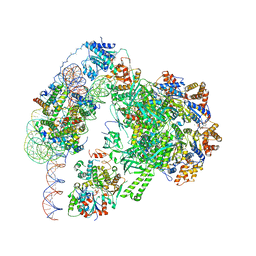 | | Chromatin remodeller-nucleosome complex at 3.6 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
6GEN
 
 | | Chromatin remodeller-nucleosome complex at 4.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
7E9F
 
 | | Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
7K7G
 
 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
6O22
 
 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
2RNW
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RSN
 
 | |
6F0Y
 
 | | Rtt109 peptide bound to Asf1 | | Descriptor: | Histone chaperone ASF1, histone acetyltransferase Rtt109 C-terminus | | Authors: | Lercher, L, Kirkpatrick, J.P, Carlomagno, T. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the Asf1-Rtt109 interaction and its role in histone acetylation.
Nucleic Acids Res., 46, 2018
|
|
2JMJ
 
 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | Descriptor: | Histone H3, Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
