1HIM
 
 | |
1HIN
 
 | |
1HIO
 
 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN, ALPHA CARBONS ONLY | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1991-09-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1HIP
 
 | | TWO-ANGSTROM CRYSTAL STRUCTURE OF OXIDIZED CHROMATIUM HIGH POTENTIAL IRON PROTEIN | | Descriptor: | HIGH POTENTIAL IRON PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Carterjunior, C.W, Kraut, J, Freer, S.T, Xuong, N.-H, Alden, R.A, Bartsch, R.G. | | Deposit date: | 1975-04-01 | | Release date: | 1976-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein.
J.Biol.Chem., 249, 1974
|
|
1HIQ
 
 | |
1HIS
 
 | |
1HIT
 
 | |
1HIV
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HIV-1 PROTEASE WITH A DIHYDROETHYLENE-CONTAINING INHIBITOR: COMPARISONS WITH MOLECULAR MODELING | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carbamoyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-1 PROTEASE | | Authors: | Thanki, N, Wlodawer, A. | | Deposit date: | 1992-02-12 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a complex of HIV-1 protease with a dihydroxyethylene-containing inhibitor: comparisons with molecular modeling.
Protein Sci., 1, 1992
|
|
1HIW
 
 | | TRIMERIC HIV-1 MATRIX PROTEIN | | Descriptor: | HIV-1 MATRIX PROTEIN, SULFATE ION | | Authors: | Hill, C.P, Worthylake, D, Bancroft, D.P, Christensen, A.M, Sundquist, W.I. | | Deposit date: | 1996-02-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1HIX
 
 | | CRYSTALLOGRAPHIC ANALYSES OF FAMILY 11 ENDO-BETA-1,4-XYLANASE XYL1 FROM STREPTOMYCES SP. S38 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Wouters, J, Georis, J, Dusart, J, Frere, J.M, Depiereux, E, Charlier, P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Analysis of Family 11 Endo-[Beta]-1,4-Xylanase Xyl1 from Streptomyces Sp. S38
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HIY
 
 | | Binding of nucleotides to NDP kinase | | Descriptor: | 3'-DEOXY 3'-AMINO ADENOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Cervoni, L, Lascu, I, Xu, Y, Gonin, P, Morr, M, Merouani, M, Janin, J, Giartoso, A. | | Deposit date: | 2001-01-05 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of Nucleotides to Nucleoside Diphosphate Kinase: A Calorimetric Study.
Biochemistry, 40, 2001
|
|
1HIZ
 
 | | Xylanase T6 (Xt6) from Bacillus Stearothermophilus | | Descriptor: | ENDO-1,4-BETA-XYLANASE, SULFATE ION, alpha-D-galactopyranose, ... | | Authors: | Sainz, G, Tepplitsky, A, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-01-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Determination of the Extracellular Xylanase from Geobacillus Stearothermophilus by Selenomethionyl MAD Phasing
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1HJ0
 
 | | Thymosin beta9 | | Descriptor: | THYMOSIN BETA9 | | Authors: | Stoll, R, Voelter, W, Holak, T.A. | | Deposit date: | 2001-01-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Conformation of Thymosin Beta9 in Water/Fluoroalcohol Solution Determined by NMR Spectroscopy
Biopolymers, 41, 1997
|
|
1HJ1
 
 | | RAT OESTROGEN RECEPTOR BETA LIGAND-BINDING DOMAIN IN COMPLEX WITH PURE ANTIOESTROGEN ICI164,384 | | Descriptor: | N-BUTYL-11-[(7R,8R,9S,13S,14S,17S)-3,17-DIHYDROXY-13-METHYL-7,8,9,11,12,13,14,15,16,17-DECAHYDRO-6H-CYCLOPENTA[A]PHENANTHREN-7-YL]-N-METHYLUNDECANAMIDE, NICKEL (II) ION, OESTROGEN RECEPTOR BETA, ... | | Authors: | Pike, A.C.W, Brzozowski, A.M, Carlquist, M. | | Deposit date: | 2001-01-08 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into the Mode of Action of a Pure Antiestrogen
Structure, 9, 2001
|
|
1HJ3
 
 | | Cytochrome cd1 Nitrite Reductase, dioxygen complex | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ4
 
 | |
1HJ5
 
 | | Cytochrome cd1 Nitrite Reductase, reoxidised enzyme | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ6
 
 | | ISOCITRATE DEHYDROGENASE S113E MUTANT COMPLEXED WITH ISOPROPYLMALATE, NADP+ AND MAGNESIUM (FLASH-COOLED) | | Descriptor: | 3-ISOPROPYLMALIC ACID, GLYCEROL, ISOCITRATE DEHYDROGENASE, ... | | Authors: | Doyle, S.A, Beernink, P.T, Koshland Junior, D.E. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Change in Substrate Specificity: Crystal Structure of S113E Isocitrate Dehydrogenase in a Complex with Isopropylmalate, Mg2+ and Nadp
Biochemistry, 40, 2001
|
|
1HJ7
 
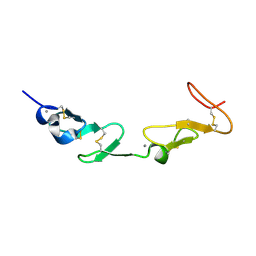 | | NMR study of a pair of LDL receptor Ca2+ binding epidermal growth factor-like domains, 20 structures | | Descriptor: | CALCIUM ION, LDL RECEPTOR | | Authors: | Saha, S, Handford, P.A, Campbell, I.D, Downing, A.K. | | Deposit date: | 2001-01-09 | | Release date: | 2001-07-11 | | Last modified: | 2018-02-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ldl Receptor Egf-Ab Pair: A Paradigm for the Assembly of Tandem Calcium Binding Egf Domains
Structure, 9, 2001
|
|
1HJ8
 
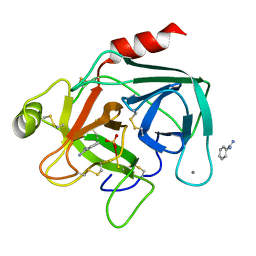 | | 1.00 AA Trypsin from Atlantic Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Leiros, H.-K.S, Mcsweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HJ9
 
 | | Atomic resolution structures of trypsin provide insight into structural radiation damage | | Descriptor: | ANILINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, McSweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-10 | | Release date: | 2002-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HJA
 
 | |
1HJB
 
 | |
1HJC
 
 | |
1HJD
 
 | |
