7BP8
 
 | | Human AAA+ ATPase VCP mutant - T76A, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BPA
 
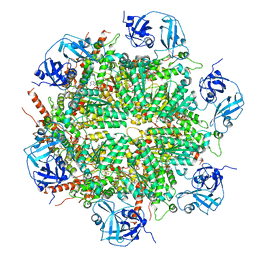 | | Human AAA+ ATPase VCP mutant - T76A, AMP-PNP-bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
1BVY
 
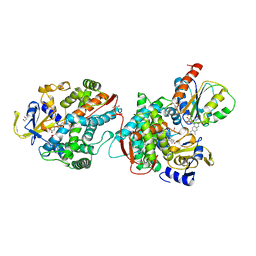 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5V7U
 
 | |
5FB7
 
 | |
5XOM
 
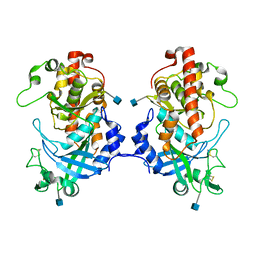 | | Hydra Fam20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosaminoglycan xylosylkinase | | Authors: | Xiao, J, Zhang, H. | | Deposit date: | 2017-05-29 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5HFJ
 
 | | crystal structure of M1.HpyAVI-SAM complex | | Descriptor: | Adenine specific DNA methyltransferase (DpnA), S-ADENOSYLMETHIONINE | | Authors: | Ma, B, Liu, W, Zhang, H. | | Deposit date: | 2016-01-07 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori
Oncotarget, 7, 2016
|
|
8UCD
 
 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | Authors: | Li, F, Bailis, J.M, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
7CQZ
 
 | | crystal structure of mouse FAM46C (TENT5C) | | Descriptor: | CHLORIDE ION, GLYCEROL, Terminal nucleotidyltransferase 5C | | Authors: | Hu, J.L, Zhang, H, Gao, S. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34969759 Å) | | Cite: | Structural and functional characterization of multiple myeloma associated cytoplasmic poly(A) polymerase FAM46C.
Cancer Commun (Lond), 41, 2021
|
|
6A8R
 
 | | Crystal structure of DUX4 HD2 domain associated with ERG DNA binding site | | Descriptor: | DNA (5'-D(P*AP*AP*TP*CP*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*TP*GP*AP*GP*AP*TP*T)-3'), Double homeobox protein 4 | | Authors: | Dong, X, Zhang, H, Cheng, N, Meng, G. | | Deposit date: | 2018-07-10 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DUX4HD2-DNAERGstructure reveals new insight into DUX4-Responsive-Element.
Leukemia, 33, 2019
|
|
6J1U
 
 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
6UVN
 
 | | CryoEM structure of VcCascasde-TniQ complex | | Descriptor: | Cas6, Cas7, Cas8/5, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type I-F CRISPR RNA guided surveillance complex bound to transposition protein TniQ.
Cell Res., 30, 2020
|
|
6V4J
 
 | | Structure of TrkH-TrkA in complex with ATP | | Descriptor: | Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
5V7R
 
 | |
3BF1
 
 | | Type III pantothenate kinase from Thermotoga maritima complexed with pantothenate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PANTOTHENOIC ACID, Type III pantothenate kinase | | Authors: | Yang, K, Huerta, C, Strauss, E, Zhang, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.
Biochemistry, 47, 2008
|
|
3BF3
 
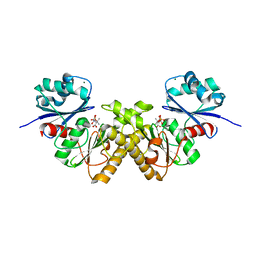 | | Type III pantothenate kinase from Thermotoga maritima complexed with product phosphopantothenate | | Descriptor: | MAGNESIUM ION, N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, Type III pantothenate kinase | | Authors: | Yang, K, Huerta, C, Strauss, E, Zhang, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.
Biochemistry, 47, 2008
|
|
3BEX
 
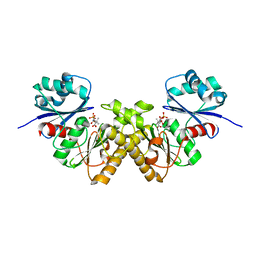 | | Type III pantothenate kinase from Thermotoga maritima complexed with pantothenate | | Descriptor: | PANTOTHENOIC ACID, PHOSPHATE ION, Type III pantothenate kinase | | Authors: | Yang, K, Huerta, C, Strauss, E, Zhang, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.
Biochemistry, 47, 2008
|
|
5ECF
 
 | |
4E5N
 
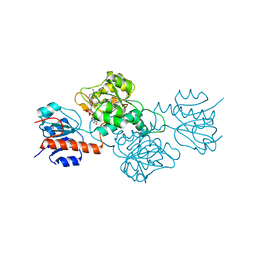 | | Thermostable phosphite dehydrogenase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Thermostable phosphite dehydrogenase | | Authors: | Zou, Y, Zhang, H, Nair, S.K. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of phosphite dehydrogenase provide insights into nicotinamide cofactor regeneration.
Biochemistry, 51, 2012
|
|
8GQ5
 
 | |
8GOT
 
 | | Crystal structure of wild-type protease 3C from Seneca Valley Virus | | Descriptor: | GLYCEROL, Peptidase C3, [(2~{S})-2-hexadecanoyloxy-3-[[(2~{R})-3-[[(2~{S})-3-[(5~{E},8~{E},11~{Z},14~{E})-icosa-5,8,11,14-tetraenoyl]oxy-2-[(9~{E},12~{Z})-octadeca-9,12-dienoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-propyl] icosanoate | | Authors: | Zhao, H.F, Zhang, H. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Allosteric regulation of Senecavirus A 3Cpro proteolytic activity by an endogenous phospholipid.
Plos Pathog., 19, 2023
|
|
8GPH
 
 | |
6V4L
 
 | | Structure of TrkH-TrkA in complex with ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
2O2F
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
