6B26
 
 | |
6B25
 
 | |
6Y4O
 
 | | Calmodulin bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-2, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83549082 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
1H71
 
 | | Psychrophilic Protease from Pseudoalteromonas 'TAC II 18' | | Descriptor: | CALCIUM ION, SERRALYSIN, ZINC ION | | Authors: | Villeret, V, Van Petegem, F, Aghajari, N, Chessa, J.-P, Gerday, C, Haser, R, Van Beeumen, J. | | Deposit date: | 2001-07-02 | | Release date: | 2003-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Psychrophilic Metalloprotease Reveal New Insights Into Catalysis by Cold-Adapted Proteases
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
6X36
 
 | | Pig R615C RyR1 in complex with CaM, EGTA (class 3, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X35
 
 | | Pig R615C RyR1 in complex with CaM, EGTA (class 1, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X33
 
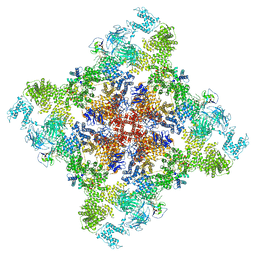 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 3, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X34
 
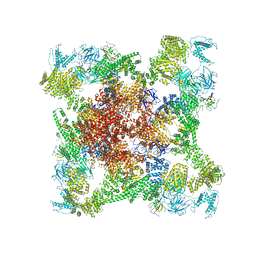 | | Pig R615C RyR1 EGTA (all classes, open) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ZINC ION | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X32
 
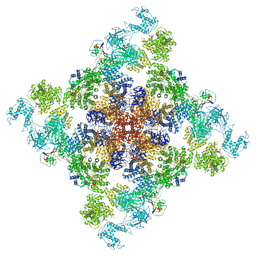 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 1 and 2, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6DAE
 
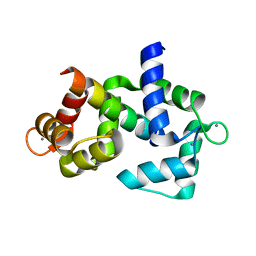 | |
6DAD
 
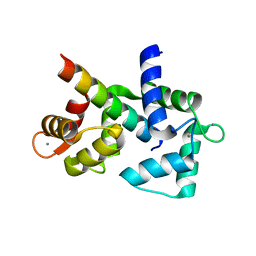 | | 1.65 Angstrom crystal structure of the N97I Ca/CaM:CaV1.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6U3D
 
 | |
6E61
 
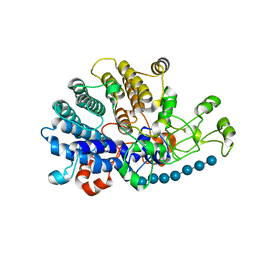 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-A in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-23 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6E9B
 
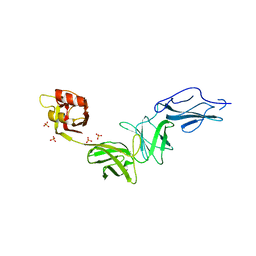 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | Mixed-linkage glucan utilization locus (MLGUL) SGBP-B, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6E60
 
 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-A | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, mixed-linkage glucan utilization locus (MLGUL) SGBP-B | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-23 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6U3A
 
 | |
6U3B
 
 | |
6U39
 
 | |
6VSV
 
 | |
6VRR
 
 | |
6UY8
 
 | |
6DAH
 
 | | 2.5 Angstrom crystal structure of the N97S CaM mutant | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6W1N
 
 | |
6ULB
 
 | |
6VHO
 
 | |
