8EFZ
 
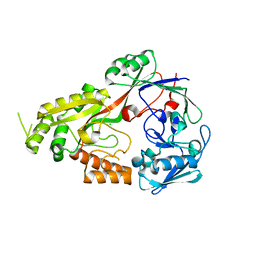 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
8F68
 
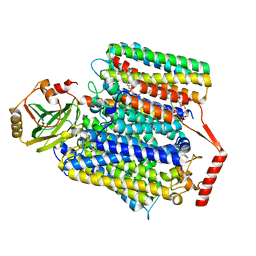 | | E. coli cytochrome bo3 ubiquinol oxidase monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
8F6C
 
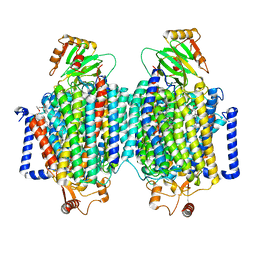 | | E. coli cytochrome bo3 ubiquinol oxidase dimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
7ROA
 
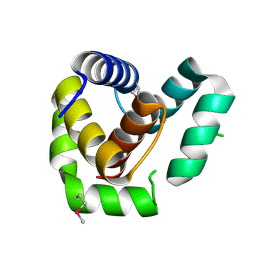 | | Crystal structure of EntV136 from Enterococcus faecalis | | Descriptor: | EntV | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-07-30 | | Release date: | 2022-10-12 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
3PP8
 
 | | 2.1 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium | | Descriptor: | Glyoxylate/hydroxypyruvate reductase A | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium.
TO BE PUBLISHED
|
|
5SUJ
 
 | | Crystal structure of uncharacterized protein LPG2148 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila.
Cell Rep, 23, 2018
|
|
5T2X
 
 | | Crystal structure of Uncharacterised protein lpg1670 | | Descriptor: | Uncharacterized protein LPG1670 | | Authors: | Chang, C, Xu, X, Cui, H, SAVCHENKO, A, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of Uncharacterised protein lpg1670
To Be Published
|
|
7LAP
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Xa | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
3KUX
 
 | | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis
To be Published
|
|
8EZR
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
3K1S
 
 | | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PTS system, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis
To be Published
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
8F09
 
 | | Crystal structure of a trimethoprim-resistant dihydrofolate reductase (DHFR) enzyme from an uncultured soil bacterium | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, D, Borek, D, Di Leo, R, Semper, C, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a trimethoprim-resistant dihydrofolate reductase (DHFR) enzyme from an uncultured soil bacterium
To Be Published
|
|
3L44
 
 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
5IBZ
 
 | | Crystal structure of a novel cyclase (pfam04199). | | Descriptor: | ACETYLPHOSPHATE, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Brown, G, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2016-02-22 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Crystal structure of a novel cyclase (pfam04199).
To Be Published
|
|
3R3S
 
 | | Structure of the YghA Oxidoreductase from Salmonella enterica | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the YghA Oxidoreductase from Salmonella enterica
TO BE PUBLISHED
|
|
3MGA
 
 | | 2.4 Angstrom Crystal Structure of Ferric Enterobactin Esterase (fes) from Salmonella typhimurium | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Enterochelin esterase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Ferric Enterobactin Esterase (fes) from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3KQF
 
 | | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Minasov, G, Halavaty, A, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-17 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis.
TO BE PUBLISHED
|
|
3CNU
 
 | | Crystal structure of the predicted coding region AF_1534 from Archaeoglobus fulgidus | | Descriptor: | Predicted coding region AF_1534 | | Authors: | Zhang, R, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the predicted coding region AF_1534 from Archaeoglobus fulgidus.
To be Published
|
|
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
1ILV
 
 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin
To Be Published
|
|
3QM3
 
 | | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fructose-bisphosphate aldolase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni
TO BE PUBLISHED
|
|
6AOJ
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal yeast Hog1p sequence | | Descriptor: | CHLORIDE ION, Ceg4, MAGNESIUM ION | | Authors: | Stogios, P.J, Nocek, B, Cuff, M.E, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
3L4E
 
 | |
