7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | 著者 | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-05-11 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
8Y87
 
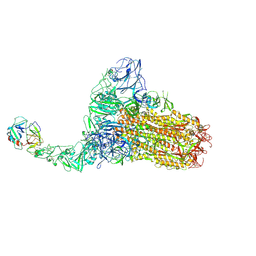 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y89
 
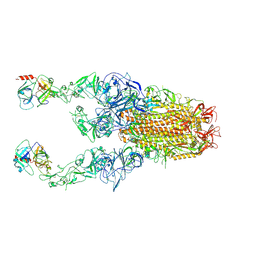 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8A
 
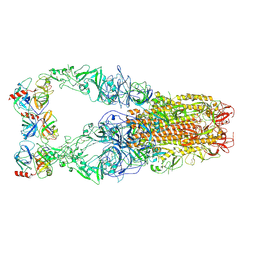 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y88
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8B
 
 | | Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Wang, H.F, Zhang, X, Lu, Y.C, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y7X
 
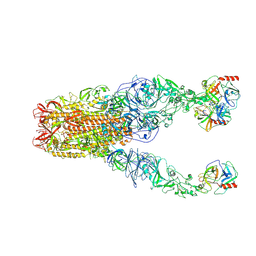 | | Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-05 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | 分子名称: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGV
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | 分子名称: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | 分子名称: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-11 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5X0Y
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | 分子名称: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | 登録日 | 2017-01-23 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.69 Å) | | 主引用文献 | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
4LGB
 
 | | ABA-mimicking ligand N-(1-METHYL-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-(4-METHYLPHENYL)METHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-1-(4-methylphenyl)methanesulfonamide, ... | | 著者 | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | 登録日 | 2013-06-27 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LG5
 
 | | ABA-mimicking ligand QUINABACTIN in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | Abscisic acid receptor PYL2, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | 著者 | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | 登録日 | 2013-06-27 | | 公開日 | 2013-08-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LGA
 
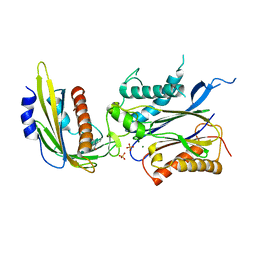 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | 著者 | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | 登録日 | 2013-06-27 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
3WB8
 
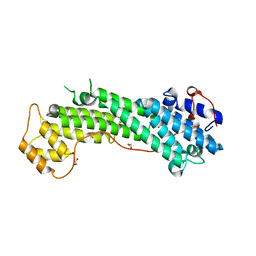 | | Crystal Structure of MyoVa-GTD | | 分子名称: | 1,2-ETHANEDIOL, Unconventional myosin-Va | | 著者 | Wei, Z, Liu, X, Yu, C, Zhang, M. | | 登録日 | 2013-05-13 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NXE
 
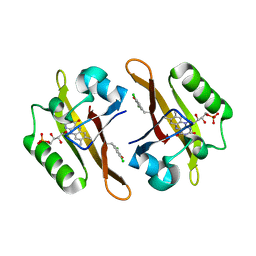 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Wang, J, Liu, X, Li, J. | | 登録日 | 2013-12-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4F5D
 
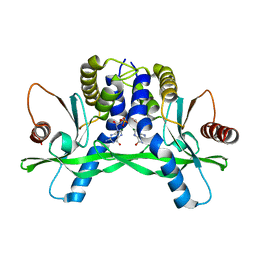 | | ERIS/STING in complex with ligand | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Transmembrane protein 173 | | 著者 | Huang, Y.H, Liu, X.Y, Su, X.D. | | 登録日 | 2012-05-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
4NXF
 
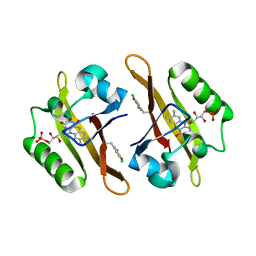 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Wang, J, Liu, X, Li, J. | | 登録日 | 2013-12-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.766 Å) | | 主引用文献 | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXG
 
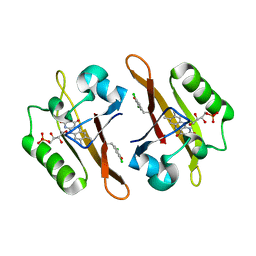 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Wang, J, Liu, X, Li, J. | | 登録日 | 2013-12-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4F5E
 
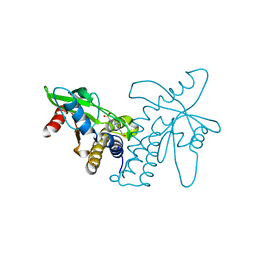 | | Crystal structure of ERIS/STING | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Transmembrane protein 173 | | 著者 | Huang, Y.H, Liu, X.Y, Su, X.D. | | 登録日 | 2012-05-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
5Y6M
 
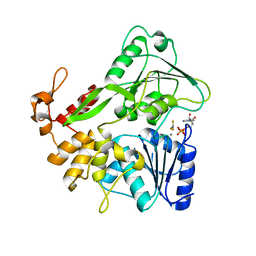 | | Zika virus helicase in complex with ADP-AlF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | 著者 | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | 登録日 | 2017-08-12 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
5UJV
 
 | | Crystal structure of FePYR1 in complex with abscisic acid | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, PYR1 | | 著者 | Ren, Z, Wang, Z, Zhou, X.E, Hong, Y, Cao, M, Chan, Z, Liu, X, Shi, H, Xu, H.E, Zhu, J.-K. | | 登録日 | 2017-01-19 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1.
Sci Rep, 7, 2017
|
|
8GY8
 
 | |
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6I
 
 | | The crystal structure of SARS-CoV-2 3C-like protease Double Mutant (L50F and E166V) in complex with a traditional Chinese Medicine Inhibitors | | 分子名称: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
