1T8Z
 
 | | Atomic Structure of A Novel Tryptophan-Zipper Pentamer | | Descriptor: | DODECAETHYLENE GLYCOL, Major outer membrane lipoprotein, SULFATE ION | | Authors: | Liu, J, Yong, W, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic structure of a tryptophan-zipper pentamer.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5F1N
 
 | | MHC complexed to 11mer peptide | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Peptide from Cytochrome P450 family 1 subfamily B polypeptide 1 | | Authors: | Liu, J, Chai, Y, Qi, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-11-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversified Anchoring Features the Peptide Presentation of DLA-88*50801: First Structural Insight into Domestic Dog MHC Class I
J Immunol., 197, 2016
|
|
4F7T
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PB1 (498-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4F7M
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
8PE9
 
 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | Deposit date: | 2023-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
6K0Y
 
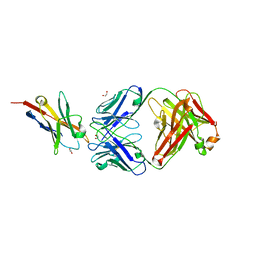 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
5H1U
 
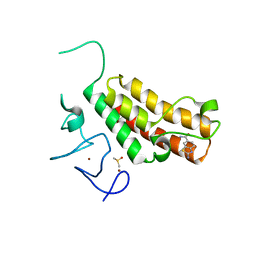 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 2 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5H1T
 
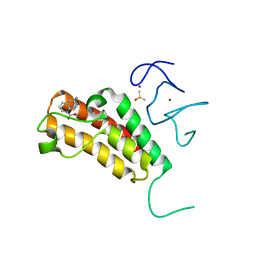 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | Descriptor: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5H1V
 
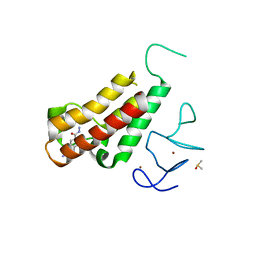 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 6 | | Descriptor: | 2-Hydrazino-1,3-benzothiazole-6-carbohydrazide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
3CK4
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, MAGNESIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
4HBK
 
 | | Structure of the Aldose Reductase from Schistosoma japonicum | | Descriptor: | Aldo-keto reductase family 1, member B4 (Aldose reductase) | | Authors: | Liu, J, Cheng, J, Zhang, X, Yang, Z, Hu, W, Xu, Y. | | Deposit date: | 2012-09-28 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aldose reductase from Schistosoma japonicum: crystallization and structure-based inhibitor screening for discovering antischistosomal lead compounds.
Parasit Vectors, 6, 2013
|
|
4F0I
 
 | | Crystal structure of apo TrkA | | Descriptor: | High affinity nerve growth factor receptor | | Authors: | Liu, J. | | Deposit date: | 2012-05-04 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Crystal Structures of TrkA and TrkB Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4U88
 
 | |
4F7P
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4FQG
 
 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | Authors: | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
3G9R
 
 | |
5H1D
 
 | | Crystal structure of C-terminal of RhoGDI2 | | Descriptor: | Rho GDP-dissociation inhibitor 2 | | Authors: | Liu, J. | | Deposit date: | 2016-10-08 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | NMR characterization of weak interactions between RhoGDI2 and fragment screening hits.
Biochim. Biophys. Acta, 1861, 2017
|
|
5HXL
 
 | |
6NE8
 
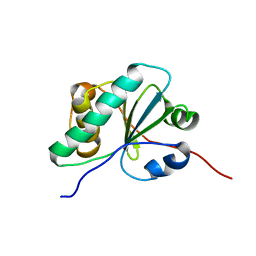 | |
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
3CRP
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-04-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
5L0O
 
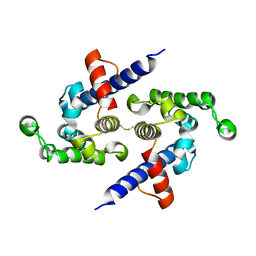 | |
7SFT
 
 | | Filamin complex-2 | | Descriptor: | Filamin-A, Integrin alpha-IIb light chain, form 2 | | Authors: | Liu, J, Qin, J. | | Deposit date: | 2021-10-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A mechanism of platelet integrin alpha IIb beta 3 outside-in signaling through a novel integrin alpha IIb subunit-filamin-actin linkage.
Blood, 141, 2023
|
|
7SC4
 
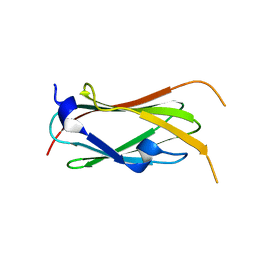 | | filamin complex-1 | | Descriptor: | Filamin-A/Integrin alpha-IIb light chain, form 2 chimera, SULFATE ION | | Authors: | Liu, J, Qin, J. | | Deposit date: | 2021-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A mechanism of platelet integrin alpha IIb beta 3 outside-in signaling through a novel integrin alpha IIb subunit-filamin-actin linkage.
Blood, 141, 2023
|
|
8I5Z
 
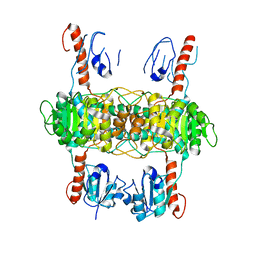 | |
