3C8X
 
 | | Crystal structure of the ligand binding domain of human Ephrin A2 (Epha2) receptor protein kinase | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3C5K
 
 | | Crystal structure of human HDAC6 zinc finger domain | | Descriptor: | Histone deacetylase 6, ZINC ION | | Authors: | Dong, A, Ravichandran, M, Schuetz, A, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human HDAC6 zinc finger domain.
To be Published
|
|
6P3A
 
 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 4-{[(3R)-1-(but-3-en-1-yl)-3-methyl-4-(oxan-4-yl)-2-oxo-1,2,3,4-tetrahydropyrido[2,3-b]pyrazin-6-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure Analysis of TAF1 Bromodomain
To Be Published
|
|
6P39
 
 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 3-methoxy-4-{[(6aR)-5-methyl-6-oxo-6,6a,7,8,9,10-hexahydro-5H-dipyrido[1,2-a:3',2'-e]pyrazin-2-yl]amino}-N-(1-methylpiperidin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Crystal Structure Analysis of TAF1 Bromodomain
To Be Published
|
|
3BRB
 
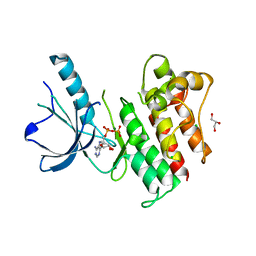 | | Crystal structure of catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
6P38
 
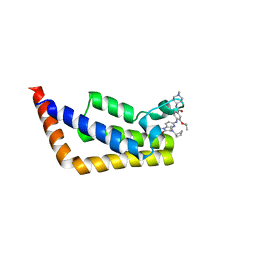 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 4-{[(3R)-4-cyclopentyl-1,3-dimethyl-2-oxo-1,2,3,4-tetrahydropyrido[2,3-b]pyrazin-6-yl]amino}-N-(1-methylpiperidin-4-yl)-3-[(propan-2-yl)oxy]benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Inhibition of TAF1 and BET Bromodomains from the BI-2536 Kinase Inhibitor Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
3BI7
 
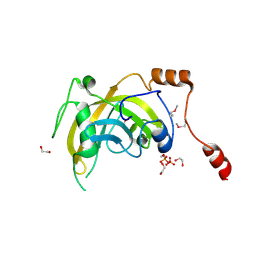 | | Crystal structure of the SRA domain of E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
3BZH
 
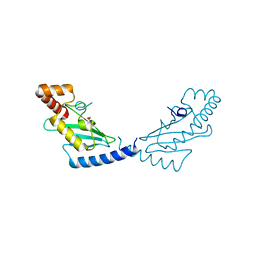 | | Crystal structure of human ubiquitin-conjugating enzyme E2 E1 | | Descriptor: | GLYCEROL, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
7MKX
 
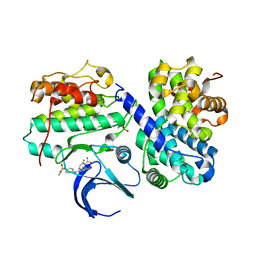 | | Crystal Structure Analysis of human CDK2 and CCNA2 complex | | Descriptor: | 2-[(5-bromo-2-{4-[(cyanomethyl)sulfamoyl]anilino}pyrimidin-4-yl)amino]-6-fluorobenzamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2021-04-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Crystal Structure Analysis of human CDK2 and CCNA2 complex
To Be Published
|
|
6OSP
 
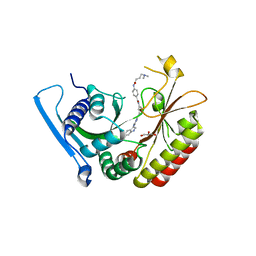 | | Crystal Structure Analysis of PIP4K2A | | Descriptor: | 4-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}-N-(3-{[6-(1H-indol-3-yl)pyrimidin-4-yl]amino}phenyl)benzamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Targeting the PI5P4K Lipid Kinase Family in Cancer Using Covalent Inhibitors.
Cell Chem Biol, 27, 2020
|
|
8EBK
 
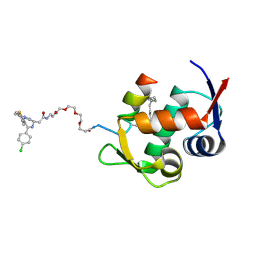 | |
8ECM
 
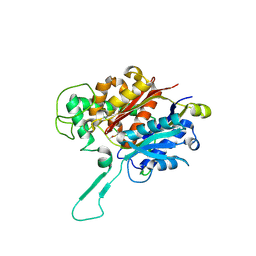 | |
6DUN
 
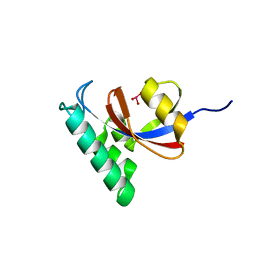 | | Crystal Structure Analysis of PIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, TRIHYDROXYARSENITE(III) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells.
Nat Commun, 9, 2018
|
|
4PYZ
 
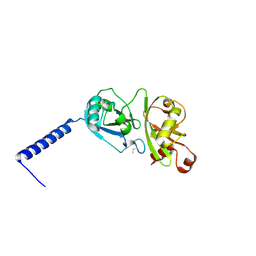 | | Crystal structure of the first two Ubl domains of Deubiquitylase USP7 | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Walker, J.R, Dong, A, Ong, M.S, Dhe-Paganon, S, Kania, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the first two Ubl domains of Deubiquitylase USP7
to be published
|
|
4WJL
 
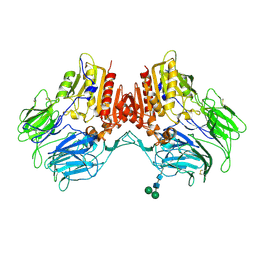 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
5K5F
 
 | | NMR structure of the HLTF HIRAN domain | | Descriptor: | Helicase-like transcription factor | | Authors: | Bezsonova, I, Neculai, D, Korzhnev, D, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HLTF HIRAN domain
To Be Published
|
|
4WVL
 
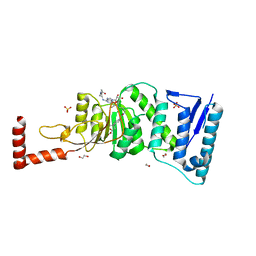 | | Structure-Guided DOT1L Probe Optimization by Label-Free Ligand Displacement | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Xu, X, Dhe-Paganon, S, Blacklow, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Guided DOT1L Probe Optimization by Label-Free Ligand Displacement.
Acs Chem.Biol., 10, 2015
|
|
6O33
 
 | | Crystal Structure Analysis of PIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, peptide | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure Analysis of PIN1
To be Published
|
|
6O34
 
 | | Crystal Structure Analysis of PIN1 | | Descriptor: | GLYCEROL, NONAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure Analysis of PIN1
To be Published
|
|
5JWM
 
 | | Bivalent BET Bromodomain Inhibition | | Descriptor: | 2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]-~{N}-[2-[2-[2-[2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethyl]ethanamide, Bromodomain-containing protein 4 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Design and characterization of bivalent BET inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
7TUO
 
 | | Crystal structure analysis of human USP28 complex with a compound | | Descriptor: | 7-amino-N-(2-{4-[(1R,3s,5S)-8-azabicyclo[3.2.1]octan-3-yl]phenyl}ethyl)-3-methylthieno[2,3-b]pyrazine-6-carboxamide, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure Analysis of human USP28 complex with a compound
To Be Published
|
|
7TUN
 
 | |
7U5W
 
 | |
7U5X
 
 | |
7SZC
 
 | |
