6LM3
 
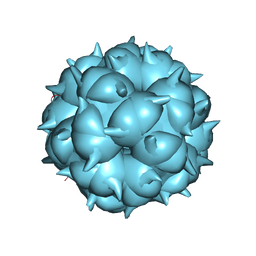 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6LPX
 
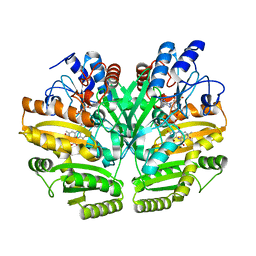 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with 2-oxoglutarate (2-OG) | | Descriptor: | 2-OXOGLUTARIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPU
 
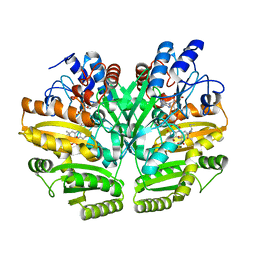 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPP
 
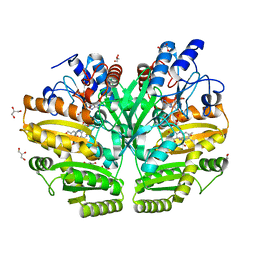 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-2-hydroxyglutarate (D-2-HG) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPQ
 
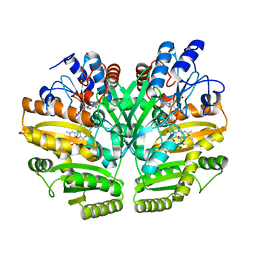 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-malate (D-MAL) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, D-MALATE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPT
 
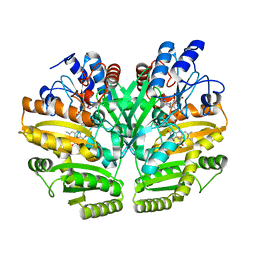 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-lactate (D-LAC) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
4XH7
 
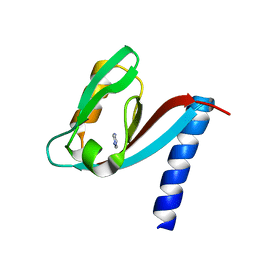 | | Crystal structure of MUPP1 PDZ4 | | Descriptor: | IMIDAZOLE, Multiple PDZ domain protein | | Authors: | Liu, Z, Zhu, H, Liu, W. | | Deposit date: | 2015-01-05 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and structural characterization of MUPP1-PDZ4 domain from Mus musculus.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
1XC1
 
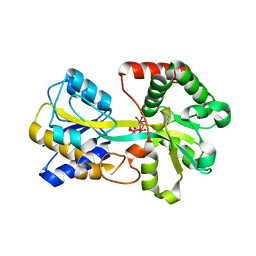 | | Oxo Zirconium(IV) Cluster in the Ferric Binding Protein (FBP) | | Descriptor: | OXO ZIRCONIUM(IV) CLUSTER, periplasmic iron-binding protein | | Authors: | Zhong, W, Alexeev, D, Harvey, I, Guo, M, Hunter, D.J.B, Zhu, H, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assembly of an Oxo-Zirconium(IV) Cluster in a Protein Cleft
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
6LPN
 
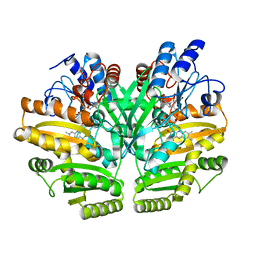 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in apo form | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
3CEY
 
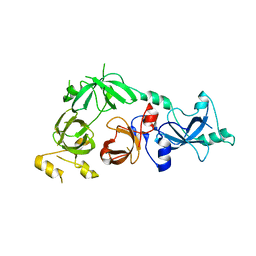 | | Crystal structure of L3MBTL2 | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein | | Authors: | Nady, N, Guo, Y, Pan, P, Allali-Hassani, A, Qi, C, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Bochkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
1CEH
 
 | | STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Kumar, A, Sekharudu, C, Ramakrishnan, B, Dupureur, C.M, Zhu, H, Tsai, M.-D, Sundaralingam, M. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water.
Protein Sci., 3, 1994
|
|
3LF5
 
 | | Structure of Human NADH cytochrome b5 oxidoreductase (Ncb5or) b5 Domain to 1.25A Resolution | | Descriptor: | Cytochrome b5 reductase 4, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Deng, B, Parthasarathy, S, Wang, W, Gibney, B.R, Battaile, K.P, Lovell, S, Benson, D.R, Zhu, H. | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Study of the individual cytochrome b5 and cytochrome b5 reductase domains of Ncb5or reveals a unique heme pocket and a possible role of the CS domain.
J.Biol.Chem., 285, 2010
|
|
6FES
 
 | | Crystal structure of novel repeat protein BRIC2 fused to DARPin D12 | | Descriptor: | 1,2-ETHANEDIOL, D12_BRIC2, a synthetic protein,D12_BRIC2, ... | | Authors: | ElGamacy, M, Coles, M, Ernst, P, Zhu, H, Hartmann, M.D, Plueckthun, A, Lupas, A. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An Interface-Driven Design Strategy Yields a Novel, Corrugated Protein Architecture.
ACS Synth Biol, 7, 2018
|
|
5K0M
 
 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
3ECQ
 
 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | Descriptor: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | Authors: | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
1EJ8
 
 | | CRYSTAL STRUCTURE OF DOMAIN 2 OF THE YEAST COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE (LYS7) AT 1.55 A RESOLUTION | | Descriptor: | CALCIUM ION, LYS7 | | Authors: | Hall, L.T, Sanchez, R.J, Holloway, S.P, Zhu, H, Stine, J.E, Lyons, T.J, Demeler, B, Schirf, V, Hansen, J.C, Nersissian, A.M, Valentine, J.S, Hart, P.J. | | Deposit date: | 2000-03-01 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic and analytical ultracentrifugation analyses of truncated and full-length yeast copper chaperones for SOD (LYS7): a dimer-dimer model of LYS7-SOD association and copper delivery.
Biochemistry, 39, 2000
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
1MOX
 
 | | Crystal Structure of Human Epidermal Growth Factor Receptor (residues 1-501) in complex with TGF-alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garrett, T.P.J, McKern, N.M, Lou, M, Elleman, T.C, Adams, T.E, Lovrecz, G.O, Zhu, H.-J, Walker, F, Frenkel, M.J, Hoyne, P.A, Jorissen, R.N, Nice, E.C, Burgess, A.W, Ward, C.W. | | Deposit date: | 2002-09-10 | | Release date: | 2003-09-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Truncated Epidermal Growth Factor Receptor Extracellular Domain Bound to Transforming Growth Factor alpha
Cell(Cambridge,Mass.), 110, 2002
|
|
6FF6
 
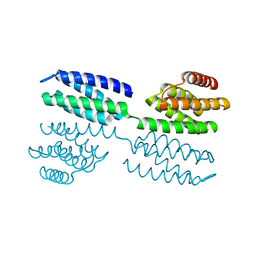 | | Crystal structure of novel repeat protein BRIC1 | | Descriptor: | BRIC1 | | Authors: | ElGamacy, M, Coles, M, Ernst, P, Zhu, H, Hartmann, M.D, Plueckthun, A, Lupas, A.N. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interface-Driven Design Strategy Yields a Novel, Corrugated Protein Architecture.
ACS Synth Biol, 7, 2018
|
|
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
8SMC
 
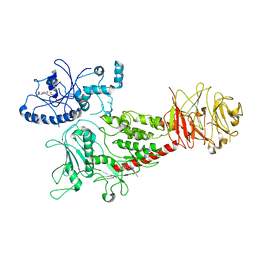 | | Cryo-EM structure of LRRK2 bound with type-I inhibitor DNL201 | | Descriptor: | 2-methyl-2-(3-methyl-4-{[4-(methylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1H-pyrazol-1-yl)propanenitrile, GUANOSINE-5'-DIPHOSPHATE, non-specific serine/threonine protein kinase | | Authors: | Sun, J, Zhu, H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Rab29-dependent asymmetrical activation of leucine-rich repeat kinase 2.
Science, 382, 2023
|
|
6LNI
 
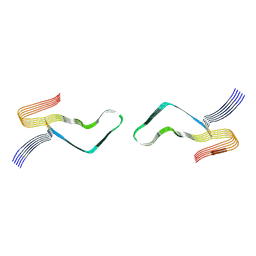 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3P4H
 
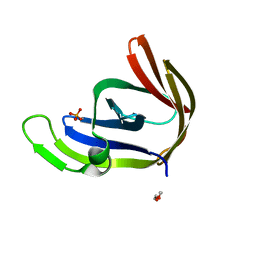 | | Structures of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Zhu, H, Shuman, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
8FH3
 
 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
