7DMC
 
 | | Dipyridamole binds to the N-terminal domain of human Hsp90A | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, CHLORIDE ION, Heat shock protein HSP 90-alpha, ... | | Authors: | Shi, L, Zhou, C, Zhong, Y, Gao, J, Zhou, H, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dipyridamole interacts with the N-terminal domain of HSP90 and antagonizes the function of the chaperone in multiple cancer cell lines.
Biochem Pharmacol, 207, 2022
|
|
7UAK
 
 | |
7WPT
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment M2-80 and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPK
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with L-Met | | Descriptor: | METHIONINE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPX
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranylimidazo[2,1-b][1,3]thiazole, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPJ
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPI
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromanyl-3-oxidanyl-phenyl)-1-[(1R)-2-[2-(2-tert-butylphenoxy)ethanoylamino]-1-(3,4-dimethoxyphenyl)ethyl]-N-methyl-benzimidazole-5-carboxamide, ACETATE ION, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WQ0
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPM
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPL
 
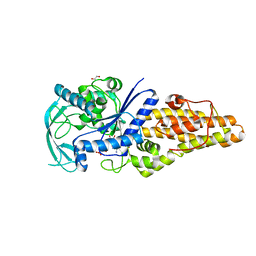 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPN
 
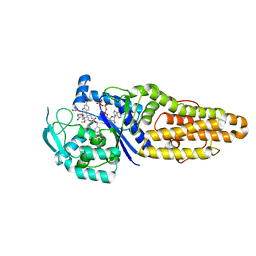 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
7C4U
 
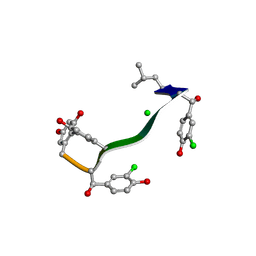 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
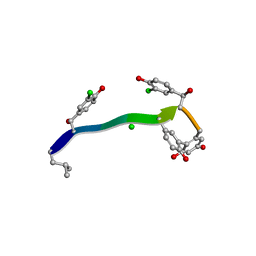 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6IDE
 
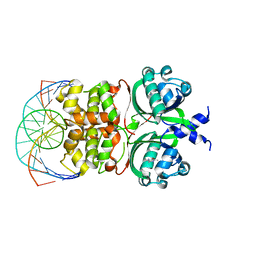 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6KJU
 
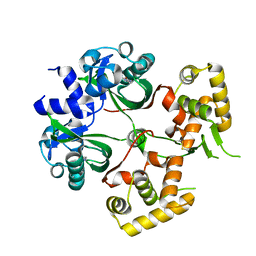 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
7C2Y
 
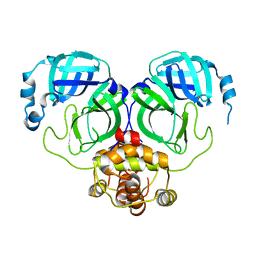 | | The crystal structure of COVID-2019 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | COVID-2019 main protease in the apo state
To Be Published
|
|
7CJY
 
 | |
7DR8
 
 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7VRB
 
 | | Structure of the Human BRG1/SS18 complex | | Descriptor: | SMARCA4 protein,Protein SSXT | | Authors: | Cheng, Y, Chen, F, Zhou, H, Long, J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Phase transition and remodeling complex assembly are important for SS18-SSX oncogenic activity in synovial sarcomas.
Nat Commun, 13, 2022
|
|
6JIO
 
 | | Human LXR-beta in complex with a ligand | | Descriptor: | Oxysterols receptor LXR-beta, tert-butyl 7-amino-3,4-dihydroisoquinoline-2(1H)-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identify liver X receptor beta modulator building blocks by developing a fluorescence polarization-based competition assay.
Eur.J.Med.Chem., 178, 2019
|
|
6K9H
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
