7EIK
 
 | | RD4-BD1 in complex with LT-872-297 | | Descriptor: | 1-[2-methyl-5-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-4-(1-methylpyrazol-4-yl)-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RD4-BD1 in complex with LT-872-297
To Be Published
|
|
7EIL
 
 | | BRD4-BD1 in complex with LT-909-110 | | Descriptor: | Bromodomain-containing protein 4, N-[4-[4-ethanoyl-5-methyl-2-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-pyrrol-3-yl]phenyl]ethanamide | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 1-(5-(1H-benzo[d]imidazole-2-yl)-2,4-dimethyl-1H-pyrrol-3-yl)ethan-1-one derivatives as novel and potent bromodomain and extra-terminal (BET) inhibitors with anticancer efficacy
Eur.J.Med.Chem., 227, 2022
|
|
7DMP
 
 | | Mouse radial spoke complex | | Descriptor: | Radial spoke head 1 homolog, Radial spoke head protein 4 homolog A, Radial spoke head protein 9 homolog | | Authors: | Zheng, W, Cong, Y. | | Deposit date: | 2020-12-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct architecture and composition of mouse axonemal radial spoke head revealed by cryo-EM
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8EHW
 
 | |
8EHX
 
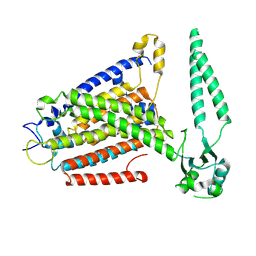 | | cryo-EM structure of TMEM63B in LMNG | | Descriptor: | CSC1-like protein 2 | | Authors: | Zheng, W, Fu, T.M, Holt, J.R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | TMEM63 proteins function as monomeric high-threshold mechanosensitive ion channels.
Neuron, 111, 2023
|
|
5WDA
 
 | | Structure of the PulG pseudopilus | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Thomassin, J.L, Bardiaux, B, Zheng, W, Nivaskumar, M, Yu, X, Nilges, M, Egelman, E.H, Izadi-Pruneyre, N, Francetic, O. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
5WCO
 
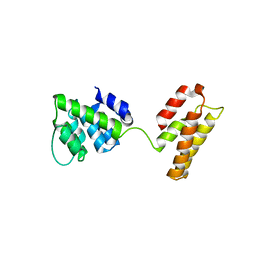 | | Matrix Protein (M1) of Infectious Salmon Anaemia Virus | | Descriptor: | NS2 | | Authors: | Zhang, W, Zheng, W, Toh, Y, Betancourt, M.A, Tu, J, Fan, Y, Vakharia, V, Liu, J, McNew, J.A, Jin, M, Tao, Y.J. | | Deposit date: | 2017-07-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of an orthomyxovirus matrix protein reveals mechanisms for self-polymerization and membrane association.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6GV9
 
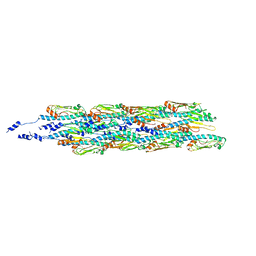 | | Structure of the type IV pilus from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Bardiaux, B, Amorim, G.C, Luna-Rico, A, Zheng, W, Guilvout, I, Jollivet, C, Nilges, M, Egelman, E, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
6WQ2
 
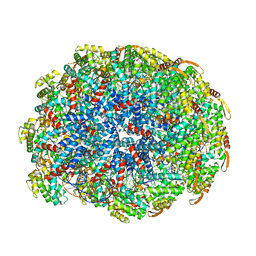 | | Cryo-EM of the S. islandicus filamentous virus, SIFV | | Descriptor: | A-DNA, Structural protein MCP1, Structural protein MCP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Zheng, W, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
5O2Y
 
 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
6DVR
 
 | | Crystal structure of human CARM1 with (R)-SKI-72 | | Descriptor: | (2R,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-methoxyphenyl)ethyl]hexanamide (non-preferred name), 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Zeng, H, Hutchinson, A, Seitova, A, Luo, M, Cai, X.C, Ke, W, Wang, J, Shi, C, Zheng, W, Lee, J.P, Ibanez, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human CARM1 with (R)-SKI-72
to be published
|
|
2CB5
 
 | | HUMAN BLEOMYCIN HYDROLASE, C73S/DELE455 MUTANT | | Descriptor: | PROTEIN (BLEOMYCIN HYDROLASE) | | Authors: | O'Farrell, P.A, Gonzalez, F, Zheng, W, Johnston, S.A, Joshua-Tor, L. | | Deposit date: | 1999-03-02 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human bleomycin hydrolase, a self-compartmentalizing cysteine protease.
Structure Fold.Des., 7, 1999
|
|
4WYB
 
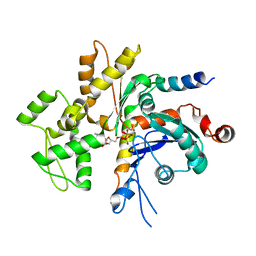 | | Structure of the Bud6 flank domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Eck, M.J, Park, E, Zheng, W. | | Deposit date: | 2014-11-17 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Structure of a Bud6/Actin Complex Reveals a Novel WH2-like Actin Monomer Recruitment Motif.
Structure, 23, 2015
|
|
5W5E
 
 | | Re-refinement of the pyocin tube structure | | Descriptor: | FIIR2 protein | | Authors: | Wang, F, Zheng, W, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
1A6R
 
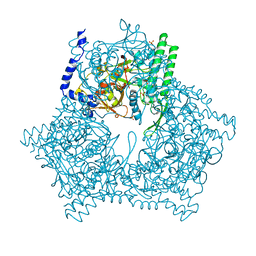 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A | | Descriptor: | GAL6, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
1CB5
 
 | | HUMAN BLEOMYCIN HYDROLASE. | | Descriptor: | BLEOMYCIN HYDROLASE | | Authors: | O'Farrell, P.A, Gonzalez, F, Zheng, W, Johnston, S.A, Joshua-Tor, L. | | Deposit date: | 1999-03-01 | | Release date: | 2000-03-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of human bleomycin hydrolase, a self-compartmentalizing cysteine protease.
Structure Fold.Des., 7, 1999
|
|
3D4B
 
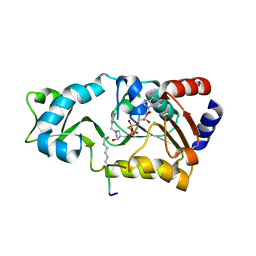 | | Crystal structure of Sir2Tm in complex with Acetyl p53 peptide and DADMe-NAD+ | | Descriptor: | 5'-O-[(R)-{[(R)-{[(3R,4R)-1-(3-carbamoylbenzyl)-4-hydroxypyrrolidin-3-yl]methoxy}(hydroxy)phosphoryl]methyl}(hydroxy)phosphoryl]adenosine, Acetyl P53 peptide, NAD-dependent deacetylase, ... | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
3D81
 
 | | Sir2-S-alkylamidate complex crystal structure | | Descriptor: | NAD-dependent deacetylase, S-alkylamidate intermediate, ZINC ION | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
3L1N
 
 | | Crystal structure of Mp1p ligand binding domain 2 complexd with palmitic acid | | Descriptor: | Cell wall antigen, PALMITIC ACID | | Authors: | Liao, S, Tung, E.T, Zheng, W, Chong, K, Xu, Y, Bartlam, M, Rao, Z, Yuen, K.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mp1p ligand binding domain 2 reveals its function as a fatty acid-binding protein.
J.Biol.Chem., 285, 2010
|
|
4FMU
 
 | | Crystal structure of Methyltransferase domain of human SET domain-containing protein 2 Compound: Pr-SNF | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-(propylamino)hexanoic acid, Histone-lysine N-methyltransferase SETD2, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zeng, H, Ibanez, G, Zheng, W, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
3GCB
 
 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A/DELTAK454 | | Descriptor: | GAL6, GLYCEROL, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
4DQM
 
 | | Revealing a marine natural product as a novel agonist for retinoic acid receptors with a unique binding mode and antitumor activity | | Descriptor: | (5S)-4-[(3E,7E)-4,8-dimethyl-10-(2,6,6-trimethylcyclohex-1-en-1-yl)deca-3,7-dien-1-yl]-5-hydroxyfuran-2(5H)-one, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Wang, S, Wang, Z, Lin, S, Zheng, W, Wang, R, Jin, S, Chen, J, Jin, L, Li, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Revealing a natural marine product as a novel agonist for retinoic acid receptors with a unique binding mode and inhibitory effects on cancer cells.
Biochem.J., 446, 2012
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
