6VGZ
 
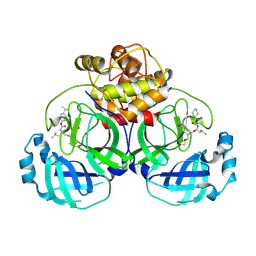 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | Descriptor: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6W2A
 
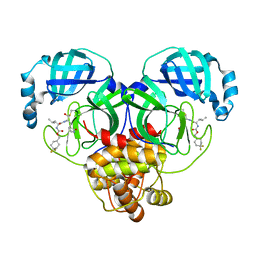 | | 1.65 A resolution structure of SARS-CoV 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a, [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
5AYW
 
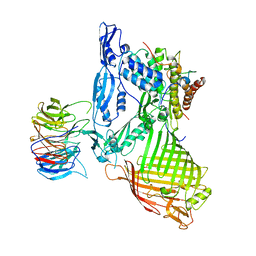 | | Structure of a membrane complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Huang, Y, Han, L, Zheng, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins
Nat.Struct.Mol.Biol., 23, 2016
|
|
3CIO
 
 | |
2KAW
 
 | | NMR structure of the mDvl1 PDZ domain in complex with its inhibitor | | Descriptor: | Segment polarity protein dishevelled homolog DVL-1, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Lee, H.J, Shao, Y, Wang, N.X, Shi, D.L, Zheng, J.J. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sulindac inhibits canonical Wnt signaling by blocking the PDZ domain of the protein Dishevelled.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2HRJ
 
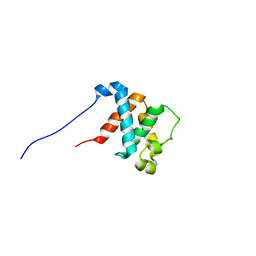 | |
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
3Q9I
 
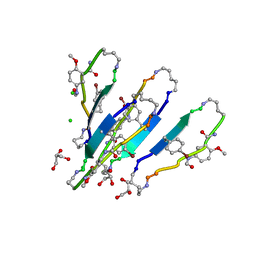 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold, bromide derivative | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide LV(4BF)FA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9G
 
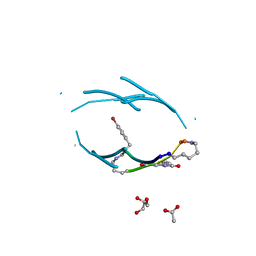 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9J
 
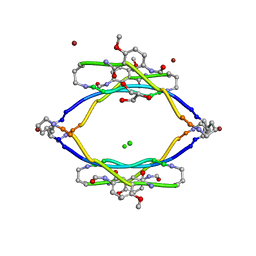 | | AIIFL segment derived from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide AIIFL(ORN)(HAO)YK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9H
 
 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | 1,4-BUTANEDIOL, Cyclic pseudo-peptide LVFFA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
2LK4
 
 | |
6LB7
 
 | | Crystal structure of the Ca2+-free and Ca2+-bound MICU1-MICU2 complex | | Descriptor: | CALCIUM ION, Calcium uptake protein 1, mitochondrial, ... | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
6LB8
 
 | | Crystal structure of the Ca2+-free T4L-MICU1-MICU2 complex | | Descriptor: | Calcium uptake protein 2, mitochondrial, Endolysin,Calcium uptake protein 1 | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
8H8C
 
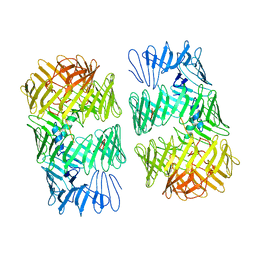 | | Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8B
 
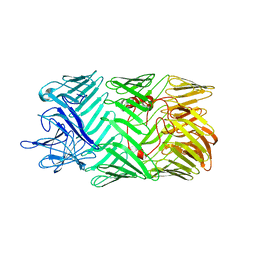 | | Type VI secretion system effector RhsP in its pre-autoproteolysis and monomeric form | | Descriptor: | Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8A
 
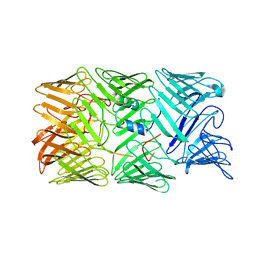 | | Type VI secretion system effector RhsP in its post-autoproteolysis and monomeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
2MAH
 
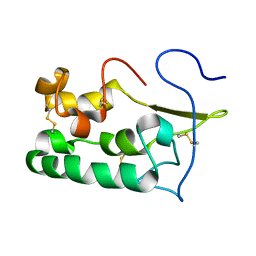 | | Solution structure of Smoothened | | Descriptor: | Protein smoothened | | Authors: | Rana, R, Lee, H, Zheng, J.J. | | Deposit date: | 2013-07-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling.
Nat Commun, 4, 2013
|
|
8IA9
 
 | |
8IAA
 
 | |
8HVR
 
 | |
2MX6
 
 | |
2L6F
 
 | | NMR Solution structure of FAT domain of FAK complexed with LD2 and LD4 motifs of PAXILLIN | | Descriptor: | Focal adhesion kinase 1, linker1, Paxillin, ... | | Authors: | Bertolucci, C.M, Guibao, C, Zhang, C, Zheng, J. | | Deposit date: | 2010-11-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Fat Domain of Fak Complexed with Ld2 and Ld4 Motifs of Paxillin
To be Published
|
|
2L6H
 
 | |
6ISL
 
 | | SnoaL-like cyclase XimE with its product xiamenmycin B | | Descriptor: | (2R,3S)-2-methyl-2-(4-methylpent-3-enyl)-3-oxidanyl-3,4-dihydrochromene-6-carboxylic acid, XimE, SnoaL-like domain protein | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
