7TWM
 
 | |
7VV9
 
 | |
7VVG
 
 | |
7VVB
 
 | |
3S29
 
 | |
5IE6
 
 | | Crystal structure of a lactonase mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE7
 
 | | Crystal structure of a lactonase double mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE5
 
 | | Crystal structure of a lactonase double mutant in complex with substrate a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5XWZ
 
 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3WGG
 
 | | Crystal structure of RSP in complex with alpha-NAD+ | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION, alpha-Diphosphopyridine nucleotide | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding mode of the oxidized alpha-anomer of NAD(+) to RSP, a Rex-family repressor
Biochem.Biophys.Res.Commun., 456, 2015
|
|
7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
8I3X
 
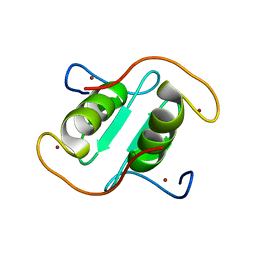 | | Rice APIP6-RING homodimer | | Descriptor: | RING-type domain-containing protein, ZINC ION | | Authors: | Zheng, Y, Zhang, X, Liu, Y, Liu, J, Wang, D. | | Deposit date: | 2023-01-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of rice APIP6 reveals a new dimerization mode of RING-type E3 ligases that facilities the construction of its working model
Phytopathol Res, 5, 2023
|
|
7EMG
 
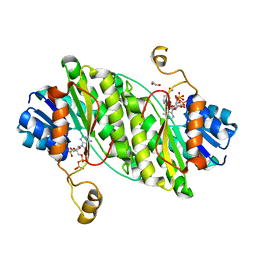 | | Carbonyl Reductase Variant 4 (R123C/L209P/F183Y/V61K) from Serratia marcescens complexed with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, Y.C, Wang, T, Bai, Y.P. | | Deposit date: | 2021-04-14 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Stereoselective synthesis of chiral delta-lactones via an engineered carbonyl reductase.
Chem.Commun.(Camb.), 57, 2021
|
|
3VMV
 
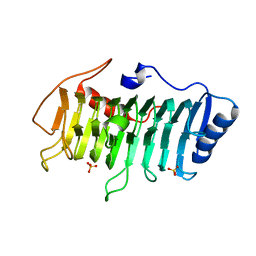 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
3VMW
 
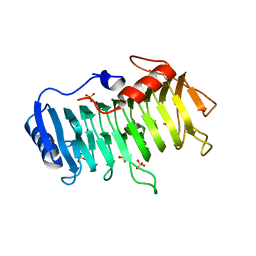 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate | | Descriptor: | Pectate lyase, SULFATE ION, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
7ULX
 
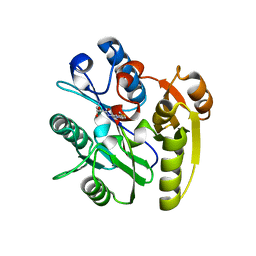 | | Human DDAH1 soaked with its inhibitor N4-(4-chloropyridin-2-yl)-L-asparagine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N-(pyridin-2-yl)-L-asparagine | | Authors: | Zheng, Y, Butrin, A, Tuley, A, Liu, D, Fast, W. | | Deposit date: | 2022-04-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Optimization of a switchable electrophile fragment into a potent and selective covalent inhibitor of human DDAH1
To Be Published
|
|
7ULV
 
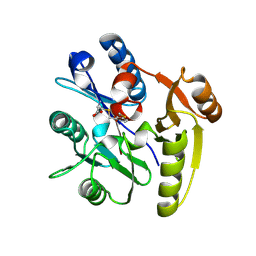 | | Human DDAH1 soaked with its inactivator S-((4-chloropyridin-2-yl)methyl)-L-cysteine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, S-[(pyridin-2-yl)methyl]-L-cysteine | | Authors: | Zheng, Y, Butrin, A, Tuley, A, Liu, D, Fast, W. | | Deposit date: | 2022-04-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Optimization of a switchable electrophile fragment into a potent and selective covalent inhibitor of human DDAH1
To Be Published
|
|
5AY7
 
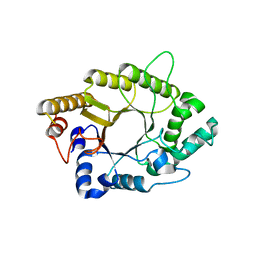 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | xylanase | | Authors: | Zheng, Y, Li, Y, Liu, W, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
5XO8
 
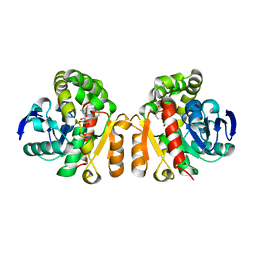 | | Crystal structure of a novel ZEN lactonase mutant with ligand Z | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5D4Y
 
 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, xylanase | | Authors: | Zheng, Y, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
1G72
 
 | | CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE HEAVY SUBUNIT, METHANOL DEHYDROGENASE LIGHT SUBUNIT, ... | | Authors: | Zheng, Y, Xia, Z, Chen, Z, Bruice, T.C, Mathews, F.S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7BWP
 
 | | Crystal complex of endo-deglycosylated PcHNL5 with (R)-mandelonitrile | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-13 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 10, 2020
|
|
5Z7J
 
 | | Crystal structure of a lactonase double mutant in complex with ligand l | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, DI(HYDROXYETHYL)ETHER, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
