6KCV
 
 | | Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide | | Descriptor: | Alginate lyase, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Dong, F, Zhang, Y.Z, Chen, X.L. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
4OHB
 
 | | Crystal structure of MilB E103A in complex with 5-hydroxymethylcytidine 5'-monophosphate (hmCMP) from Streptomyces rimofaciens | | Descriptor: | 5-(hydroxymethyl)cytidine 5'-(dihydrogen phosphate), CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
4HHE
 
 | | Quinolinate synthase from Pyrococcus furiosus | | Descriptor: | CHLORIDE ION, Quinolinate synthase A | | Authors: | Soriano, E.V, Zhang, Y, Settembre, E.C, Colabroy, K, Sanders, J.M, Dorrestein, P.C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-10-09 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Active-site models for complexes of quinolinate synthase with substrates and intermediates.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6L4R
 
 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
6BQ1
 
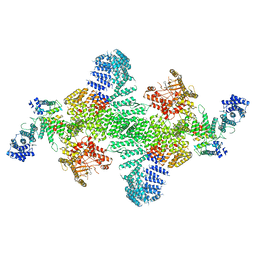 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4KYS
 
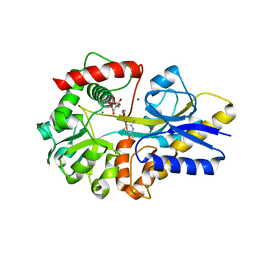 | | Clostridium botulinum thiaminase I in complex with thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Sikowitz, M.D, Shome, B, Zhang, Y, Begley, T.P, Ealick, S.E. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure of a Clostridium botulinum C143S Thiaminase I/Thiamin Complex Reveals Active Site Architecture.
Biochemistry, 52, 2013
|
|
6NKO
 
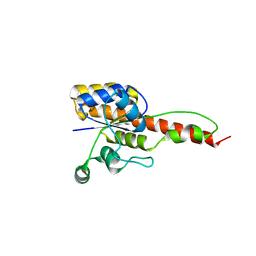 | | Crystal structure of ForH | | Descriptor: | ForH | | Authors: | Zheng, J, Irani, S, Zhang, Y. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Identification of the Formycin A Biosynthetic Gene Cluster from Streptomyces kaniharaensis Illustrates the Interplay between Biological Pyrazolopyrimidine Formation and de Novo Purine Biosynthesis.
J. Am. Chem. Soc., 141, 2019
|
|
4RM5
 
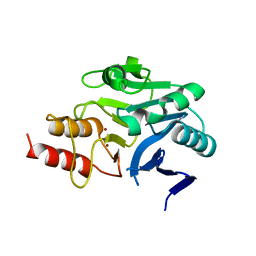 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4S29
 
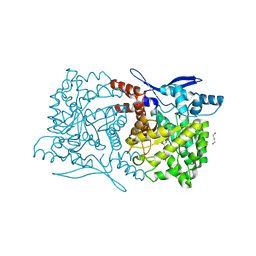 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide and Fe | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, FE (II) ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S27
 
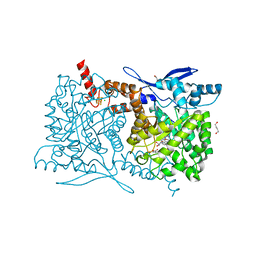 | | Crystal structure of Arabidopsis thaliana ThiC with bound aminoimidazole ribonucleotide, 5'-deoxyadenosine, L-methionine, Fe4S4 cluster and Fe | | Descriptor: | 1,4-BUTANEDIOL, 5'-DEOXYADENOSINE, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S25
 
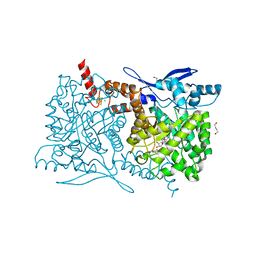 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (trigonal crystal form) | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S2A
 
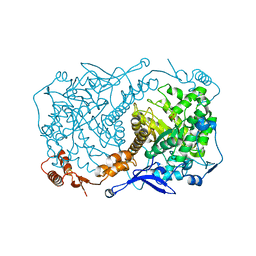 | | Crystal structure of Caulobacter crescentus ThiC with Fe4S4 cluster at remote site (holo form) | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, Phosphomethylpyrimidine synthase | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S28
 
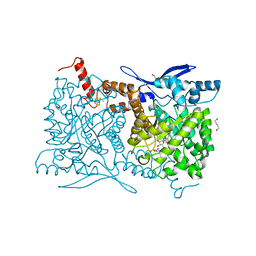 | | Crystal structure of Arabidopsis thaliana ThiC with bound aminoimidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Fe | | Descriptor: | 1,4-BUTANEDIOL, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
5XDZ
 
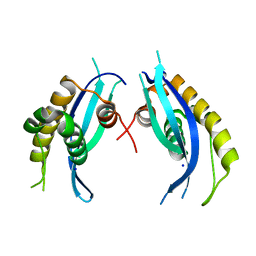 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
5TOU
 
 | | STRUCTURE OF C-PHYCOCYANIN FROM ARCTIC PSEUDANABAENA SP. LW0831 | | Descriptor: | PHYCOCYANOBILIN, Phycocyanin alpha-1 subunit, Phycocyanin beta-1 subunit | | Authors: | Wang, Q.M, Li, C.Y, Su, H.N, Zhang, Y.Z, Xie, B.B. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the cold adaptation of the photosynthetic pigment-protein C-phycocyanin from an Arctic cyanobacterium
Biochim. Biophys. Acta, 1858, 2017
|
|
5YIU
 
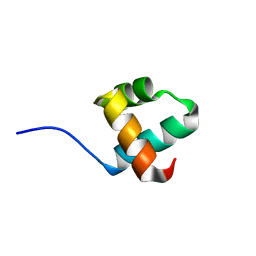 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) | | Descriptor: | Cell cycle regulatory protein GcrA | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIW
 
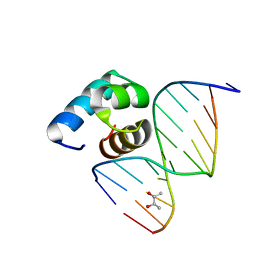 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) in complex with methylated dsDNA (crystal form 2) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*(6MA)P*TP*TP*CP*GP*C)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIV
 
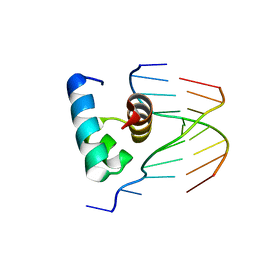 | |
4QC2
 
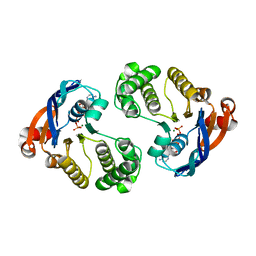 | | Crystal structure of lipopolysaccharide transport protein LptB in complex with ATP and Magnesium ions | | Descriptor: | ABC transporter related protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, Z, Xiang, Q, Zhu, X, Dong, H, He, C, Wang, H, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
5YIX
 
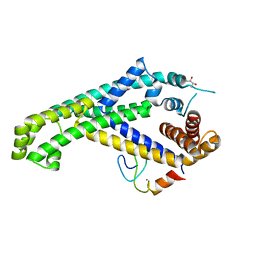 | | Caulobacter crescentus GcrA sigma-interacting domain (SID) in complex with domain 2 of sigma 70 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, RNA polymerase sigma factor RpoD, ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
6VUA
 
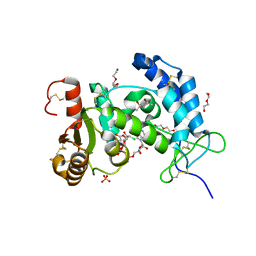 | | X-ray structure of human CD38 catalytic domain with 2'-Cl-araNAD+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, Z, Zhang, X.N, Nasertorabi, F, Han, G.W, Stevens, R.C, Zhang, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis of site-specific antibody-drug conjugates by ADP-ribosyl cyclases.
Sci Adv, 6, 2020
|
|
7TCL
 
 | | Crystal structure of P.IsnB complexed with tyrosine isonitrile | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-2-isocyanopropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-12-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Elucidation of divergent desaturation pathways in the formation of vinyl isonitrile and isocyanoacrylate.
Nat Commun, 13, 2022
|
|
5Z7I
 
 | | Caulobacter crescentus GcrA DNA-binding domain(DBD)in complex with unmethylated dsDNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*AP*TP*TP*CP*GP*C*)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
6KBW
 
 | | Crystal structure of Tmm from Myroides profundi D25 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethylamine monooxygenase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Oxidation of trimethylamine to trimethylamine N-oxide facilitates high hydrostatic pressure tolerance in a generalist bacterial lineage.
Sci Adv, 7, 2021
|
|
4S2S
 
 | | Crystal Structure of Fab fragment of monoclonal antibody RoAb13 | | Descriptor: | RoAb13 Fab Heavy chain, RoAb13 Fab Light chain | | Authors: | Chain, B, Arnold, J, Akthar, S, Noursadeghi, M, Lapp, T, Ji, C, Naider, D, Zhang, Y, Govada, L, Saridakis, E, Chayen, N.E. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Linear Epitope in the N-Terminal Domain of CCR5 and Its Interaction with Antibody.
Plos One, 10, 2015
|
|
