5H93
 
 | |
5H98
 
 | |
4E55
 
 | |
4E56
 
 | |
6O2P
 
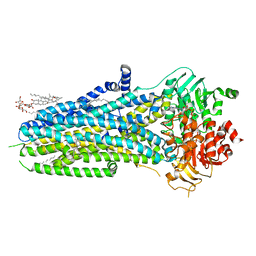 | | Complex of ivacaftor with cystic fibrosis transmembrane conductance regulator (CFTR) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | 著者 | Liu, F, Zhang, Z, Chen, J, Levit, A, Shoichet, B. | | 登録日 | 2019-02-24 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural identification of a hotspot on CFTR for potentiation.
Science, 364, 2019
|
|
3GBU
 
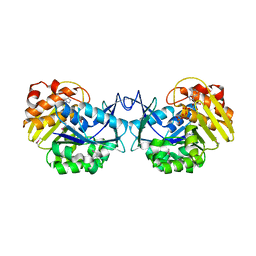 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized sugar kinase PH1459 | | 著者 | Eswaramoorthy, S, Kumar, G, Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-20 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with ATP
To be Published
|
|
3L72
 
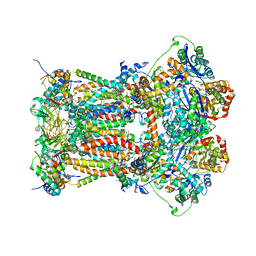 | | Chicken cytochrome BC1 complex with kresoxim-I-dimethyl bound | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME B, ... | | 著者 | Huang, L, Zhang, Z, Berry, E.A. | | 登録日 | 2009-12-27 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "DISTAL" Qo inhibitors.
To be Published
|
|
5VRF
 
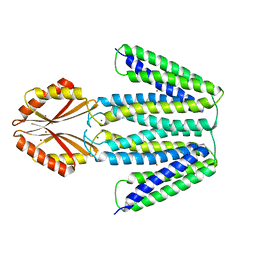 | | CryoEM Structure of the Zinc Transporter YiiP from helical crystals | | 分子名称: | Cadmium and zinc efflux pump FieF, ZINC ION | | 著者 | Coudray, N, Lopez-Redondo, M, Zhang, Z, Alexopoulos, J, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | 登録日 | 2017-05-10 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8GS9
 
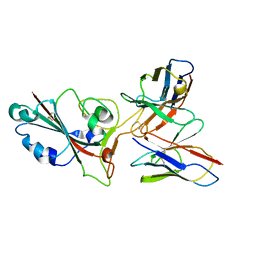 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | 分子名称: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | 著者 | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | 登録日 | 2022-09-05 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
8GSB
 
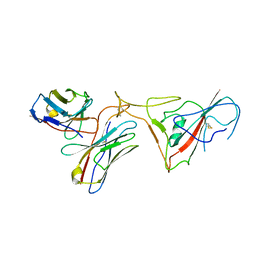 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
2MNG
 
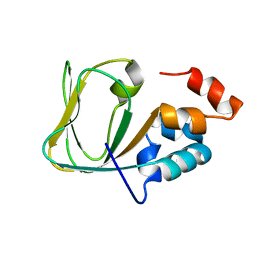 | | Apo Structure of human HCN4 CNBD solved by NMR | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | 登録日 | 2014-04-03 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
2OEH
 
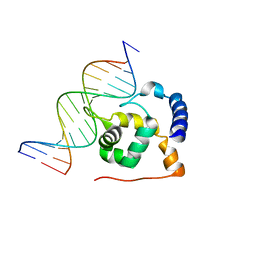 | |
5UAK
 
 | |
5SV9
 
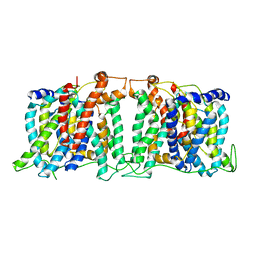 | | Structure of the SLC4 transporter Bor1p in an inward-facing conformation | | 分子名称: | Bor1p boron transporter | | 著者 | Coudray, N, Seyler, S, Lasala, R, Zhang, Z, Clark, K.M, Dumont, M.E, Rohou, A, Beckstein, O, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | 登録日 | 2016-08-05 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Structure of the SLC4 transporter Bor1p in an inward-facing conformation.
Protein Sci., 26, 2017
|
|
2QCJ
 
 | | Native Structure of Lyp | | 分子名称: | Tyrosine-protein phosphatase non-receptor type 22 | | 著者 | Sun, J.P, Yu, X, Zhang, Z.Y. | | 登録日 | 2007-06-19 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of Lyp and its complex with a selective inhibitor
To be Published
|
|
2QCT
 
 | | Structure of Lyp with inhibitor I-C11 | | 分子名称: | 1,2-ETHANEDIOL, 6-HYDROXY-3-{(4R)-1-[4-(1-NAPHTHYLAMINO)-4-OXOBUTYL]-1,2,3-TRIAZOLIDIN-4-YL}-1-BENZOFURAN-5-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 22 | | 著者 | Yu, X, Sun, J.P, Zhang, Z.Y. | | 登録日 | 2007-06-19 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Lyp and its complex with a selective inhibitor
To be Published
|
|
2QMZ
 
 | |
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | 分子名称: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | 分子名称: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2019-03-30 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | 分子名称: | Glycoprotein, scFv 523-11 | | 著者 | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | 登録日 | 2019-12-06 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9037 Å) | | 主引用文献 | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | 分子名称: | Glycoprotein,Glycoprotein,Glycoprotein | | 著者 | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | 登録日 | 2019-12-06 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.097 Å) | | 主引用文献 | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | 分子名称: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | 著者 | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | 登録日 | 2021-08-25 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | 分子名称: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | 著者 | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | 登録日 | 2021-08-25 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
