6E9E
 
 | | EsCas13d-crRNA binary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, crRNA (52-MER) | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
6E9F
 
 | | EsCas13d-crRNA-target RNA ternary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, RNA (27-MER), ... | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
8YA5
 
 | |
7BTA
 
 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTD
 
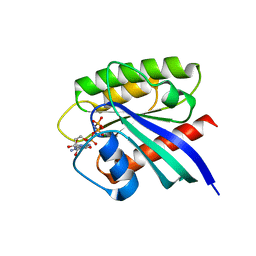 | | Crystal structure of Rheb Y35N mutant bound to GppNHp | | Descriptor: | GTP-binding protein Rheb, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
8YA1
 
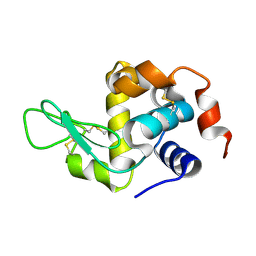 | | HEN EGG WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8YA4
 
 | |
8Y9W
 
 | |
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
8Y9V
 
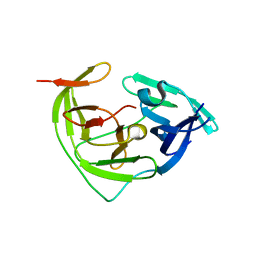 | | ZIKV NS2B/NS3 protease | | Descriptor: | DAR-LYS-ORN-ARG, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H, Wang, Q.S. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
6L7P
 
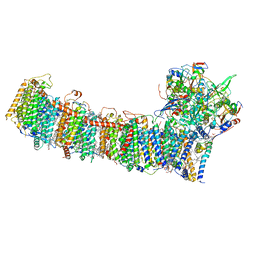 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
7BTC
 
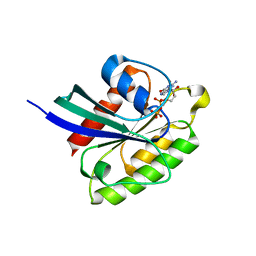 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
8J8D
 
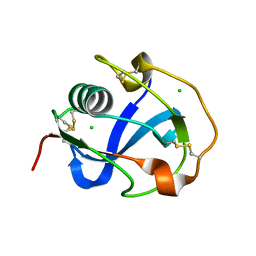 | |
8J8T
 
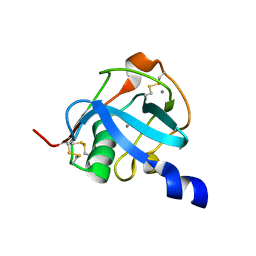 | |
8W6P
 
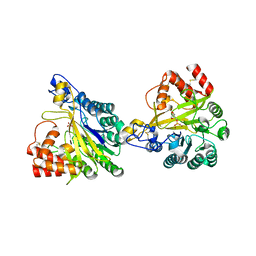 | | Crystal structure of dimeric murine SMPDL3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S, Hou, Y. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | Descriptor: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
2DHO
 
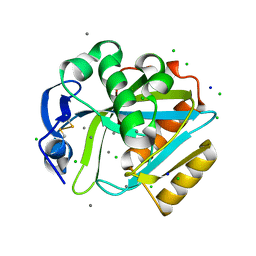 | | Crystal structure of human IPP isomerase I in space group P212121 | | Descriptor: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-03-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
8W6R
 
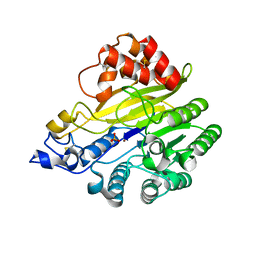 | | murine SMPDL3A bound to sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
7BXX
 
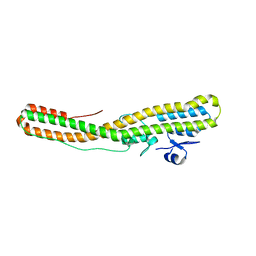 | | Tetanus neurotoxin translocation domain -C467S | | Descriptor: | Tetanus toxin | | Authors: | Zhang, C.M, Imoto, Y, Fukuda, Y, Yamashita, E, Inoue, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | SOLUTION SCATTERING (2.34 Å), X-RAY DIFFRACTION | | Cite: | Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X, 5, 2021
|
|
7BY4
 
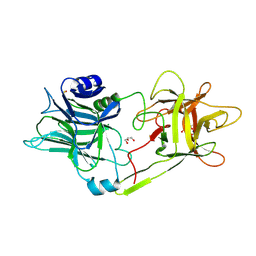 | | Tetanus neurotoxin receptor binding domain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Zhang, C.M, Imoto, Y, Inoue, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X, 5, 2021
|
|
7BY5
 
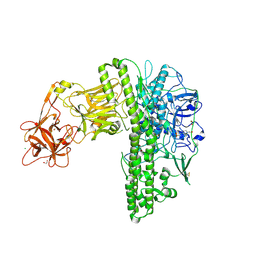 | | Tetanus neurotoxin mutant-(H233A/E234Q/H237A/Y375F) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Zhang, C.M, Imoto, Y, Fukuda, Y, Yamashita, E, Inoue, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X, 5, 2021
|
|
7BYY
 
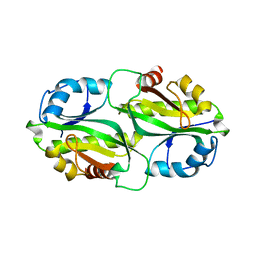 | | Crystal structure of bacterial toxin | | Descriptor: | Acetyltransferase | | Authors: | Zhang, C, Yashiro, Y, Tomita, K. | | Deposit date: | 2020-04-25 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Substrate specificities of Escherichia coli ItaT that acetylates aminoacyl-tRNAs.
Nucleic Acids Res., 48, 2020
|
|
6KIK
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | Descriptor: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIY
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KY6
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | Descriptor: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
