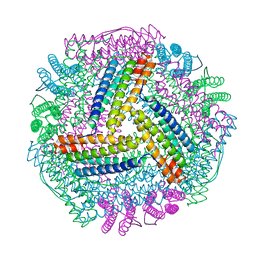
8IQV
 
 | |
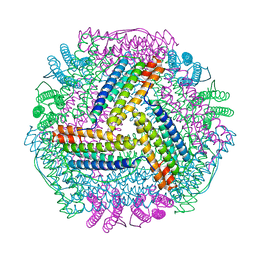
8IQY
 
 | |
7L13
 
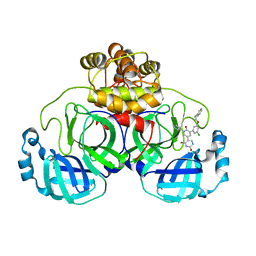 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
1YXT
 
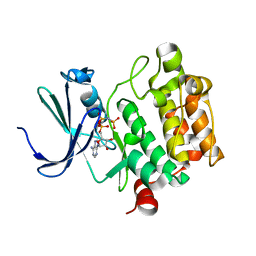 | | Crystal Structure of Kinase Pim1 in complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of proto-oncogene kinase Pim1: a target of aberrant somatic hypermutations in diffuse large cell lymphoma.
J.Mol.Biol., 348, 2005
|
|
6EOA
 
 | | Crystal Structure of HAL3 from Cryptococcus neoformans | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phosphopantothenoylcysteine decarboxylase | | Authors: | Reverter, D, Zhang, C, Molero, C, Arino, J. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Characterization of the atypical Ppz/Hal3 phosphatase system from the pathogenic fungus Cryptococcus neoformans.
Mol.Microbiol., 111, 2019
|
|
1YXS
 
 | | Crystal Structure of Kinase Pim1 with P123M mutation | | Descriptor: | IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of proto-oncogene kinase Pim1: a target of aberrant somatic hypermutations in diffuse large cell lymphoma.
J.Mol.Biol., 348, 2005
|
|
4RT7
 
 | | Crystal Structure of FLT3 with a small molecule inhibitor | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397.
Cancer Discov, 5, 2015
|
|
5Z9I
 
 | |
2O63
 
 | | Crystal structure of Pim1 with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
1YXV
 
 | | Crystal Structure of Kinase Pim1 in complex with 3,4-Dihydroxy-1-methylquinolin-2(1H)-one | | Descriptor: | 3,4-DIHYDROXY-1-METHYLQUINOLIN-2(1H)-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Proto-oncogene Kinase Pim1: A Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma.
J.Mol.Biol., 348, 2005
|
|
1YWV
 
 | | Crystal Structures of Proto-Oncogene Kinase Pim1: a Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma | | Descriptor: | IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of proto-oncogene kinase Pim1: a target of aberrant somatic hypermutations in diffuse large cell lymphoma.
J.Mol.Biol., 348, 2005
|
|
7T1F
 
 | | Crystal structure of GDP-bound T50I mutant of human KRAS4B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analyses of a germline KRAS T50I mutation provide insights into Raf activation.
JCI Insight, 8, 2023
|
|
3UON
 
 | | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, CHLORIDE ION, Human M2 muscarinic acetylcholine, ... | | Authors: | Haga, K, Kruse, A.C, Asada, H, Yurugi-Kobayashi, T, Shiroishi, M, Zhang, C, Weis, W.I, Okada, T, Kobilka, B.K, Haga, T, Kobayashi, T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.
Nature, 482, 2012
|
|
3R5S
 
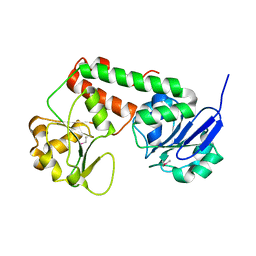 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
6PT0
 
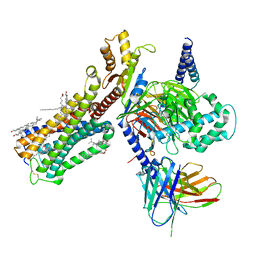 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
8W72
 
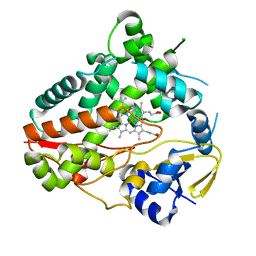 | |
4Q79
 
 | | Structure of a HG-derivative CsgG | | Descriptor: | CsgG, MERCURY (II) ION | | Authors: | Huang, Y, Zhang, C.X, Cao, B, Zhao, Y, Kou, Y, Ni, D. | | Deposit date: | 2014-04-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the nonameric bacterial amyloid secretion channel
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8IQX
 
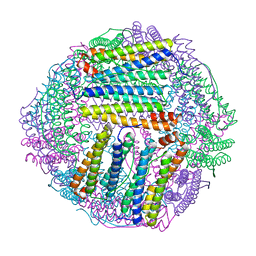 | | ferritin mutant-P156H | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
7M3G
 
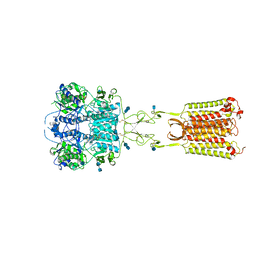 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
3FCU
 
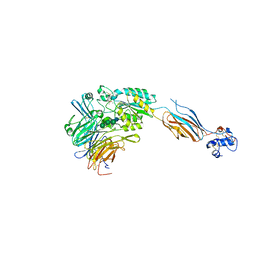 | | Structure of headpiece of integrin aIIBb3 in open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
8JNQ
 
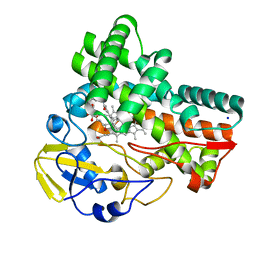 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JOO
 
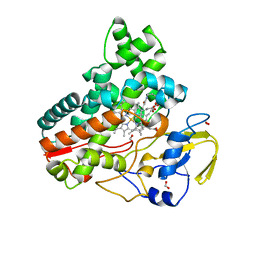 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JUA
 
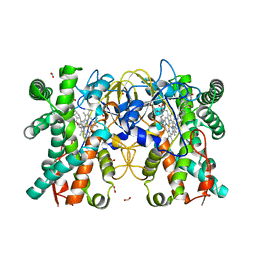 | | Multifunctional cytochrome P450 enzyme IkaD from Streptomyces sp. ZJ306, in complex with epoxyikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,13R,15S,16R,17S,19Z,26S)-11-ethyl-2-hydroxy-10-methyl-22,27-diaza-14 oxahexacyclo[24.2.1.05,17.07,16.013,15.08,12]nonacosa-1(2),3,19-triene-21,28,29-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.00001121 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2X4Z
 
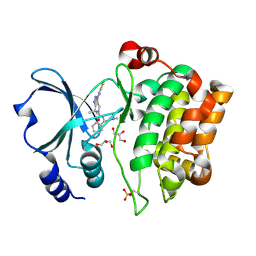 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with PF-03758309 | | Descriptor: | GLYCEROL, PF-3758309, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.R, Deng, Y, Murray, B, Guo, C, Piraino, J, Westwick, J, Zhang, C, Lamerdin, J, Dagostino, E, Loi, C.-M, Zager, M, Kraynov, E, Christensen, J, Martinez, R, Kephart, S, Marakovits, J, Karlicek, S, Bergqvist, S, Smeal, T. | | Deposit date: | 2010-02-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-Molecule P21-Activated Kinase Inhibitor Pf- 3758309 is a Potent Inhibitor of Oncogenic Signaling and Tumor Growth.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8HGH
 
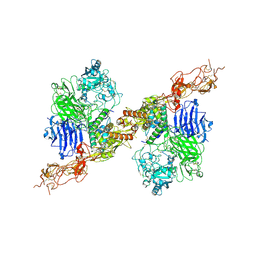 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
