8OVX
 
 | | Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CTF19, Inner kinetochore subunit MCM21, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
6KG3
 
 | |
6KH2
 
 | |
4E55
 
 | |
2OEH
 
 | |
8A5Y
 
 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
8A61
 
 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
8A3T
 
 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
8HPA
 
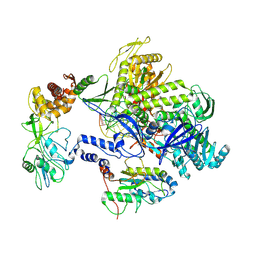 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-31 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
5BX1
 
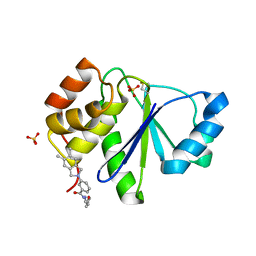 | | Crystal Structure of PRL-1 complex with compound analogy 3 | | Descriptor: | 3-(5,6-dimethyl-2H-isoindol-2-yl)-N'-[(E)-furan-2-ylmethylidene]benzohydrazide, Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Liu, S, Bai, Y, Zhang, Z. | | Deposit date: | 2015-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of PRL-1 complex with compound analogy 3
To Be Published
|
|
4E56
 
 | |
6U8V
 
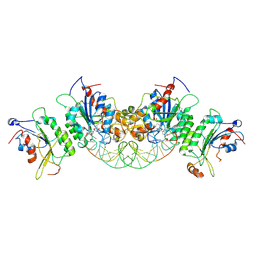 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
4H5S
 
 | | Complex structure of Necl-2 and CRTAM | | Descriptor: | Cell adhesion molecule 1, Cytotoxic and regulatory T-cell molecule | | Authors: | Zhang, S, Lu, G, Qi, J, Li, Y, Zhang, Z, Zhang, B, Yan, J, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Competition of cell adhesion and immune recognition: insights into the interaction between CRTAM and nectin-like 2.
Structure, 21, 2013
|
|
6U8W
 
 | | Crystal structure of DNMT3B(K777A)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.94891548 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
5A31
 
 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
3ZZM
 
 | | Crystal structure of Mycobacterium tuberculosis PurH with a novel bound nucleotide CFAIR, at 2.2 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, GLYCEROL, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-02 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
2QCT
 
 | | Structure of Lyp with inhibitor I-C11 | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXY-3-{(4R)-1-[4-(1-NAPHTHYLAMINO)-4-OXOBUTYL]-1,2,3-TRIAZOLIDIN-4-YL}-1-BENZOFURAN-5-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Yu, X, Sun, J.P, Zhang, Z.Y. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Lyp and its complex with a selective inhibitor
To be Published
|
|
4AEZ
 
 | | Crystal Structure of Mitotic Checkpoint Complex | | Descriptor: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | Authors: | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
4A2N
 
 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
2QMZ
 
 | |
1CFM
 
 | | CYTOCHROME F FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chi, Y.I, Huang, L.S, Zhang, Z, Fernandez-Velasco, J.G, Malkin, R, Berry, E.A. | | Deposit date: | 1998-09-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a truncated form of cytochrome f from chlamydomonas reinhardtii.
Biochemistry, 39, 2000
|
|
4BH6
 
 | | Insights into degron recognition by APC coactivators from the structure of an Acm1-Cdh1 complex | | Descriptor: | APC/C ACTIVATOR PROTEIN CDH1, APC/C-CDH1 MODULATOR 1 | | Authors: | He, J, Chao, W.C.H, Zhang, Z, Yang, J, Cronin, N, Barford, D. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Degron Recognition by Apc/C Coactivators from the Structure of an Acm1-Cdh1 Complex.
Mol.Cell, 50, 2013
|
|
