4LRM
 
 | | EGFR D770_N771insNPG in complex with PD168393 | | Descriptor: | Epidermal growth factor receptor, N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2013-07-20 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer.
Sci Transl Med, 5, 2013
|
|
4LQM
 
 | | EGFR L858R in complex with PD168393 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[4-(3-BROMO-PHENYLAMINO)-QUINAZOLIN-6-YL]-ACRYLAMIDE | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2013-07-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer.
Sci Transl Med, 5, 2013
|
|
7BTT
 
 | | A X-ray cocrystal structure of XMU-MP-5 bound to the ALK kinase domain | | Descriptor: | 2-(dimethylamino)-1-[5-methoxy-6-[[4-[(2-propan-2-ylsulfonylphenyl)amino]-5H-pyrrolo[3,2-d]pyrimidin-2-yl]amino]-2,3-dihydroindol-1-yl]ethanone, ALK tyrosine kinase receptor | | Authors: | Yun, C.H, Zhu, S.J. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A X-ray cocrystal structure of XMU-MP-5 bound to the ALK kinase domain
To Be Published
|
|
4WD5
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with QL-X138 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-{2-methyl-5-[2-oxo-9-(1H-pyrazol-4-yl)benzo[h][1,6]naphthyridin-1(2H)-yl]phenyl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2014-09-07 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of EGFR 696-1022 T790M in complex with QL-X138
To Be Published
|
|
6P8Q
 
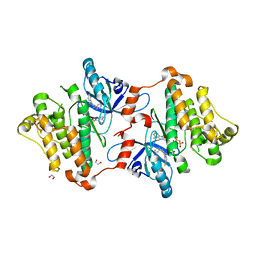 | | EGFR in complex with a dihydrodibenzodiazepinone allosteric inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 10-benzyl-2-fluoro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Yun, C.H, Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Dibenzodiazepinones as Allosteric Mutant-Selective EGFR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5X2A
 
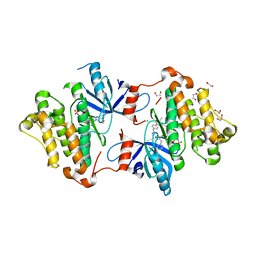 | |
5X27
 
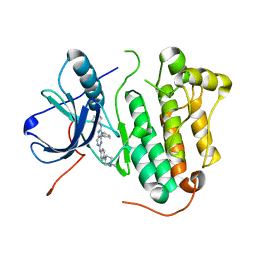 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X28
 
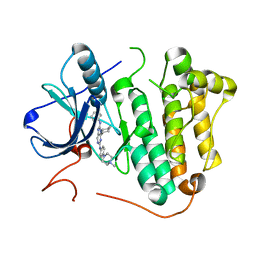 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2C
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(5) | | Descriptor: | 1,2-ETHANEDIOL, 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, ... | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2F
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
6JXT
 
 | |
6JX0
 
 | |
6JWL
 
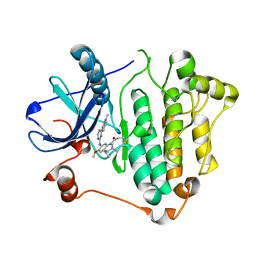 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
5J87
 
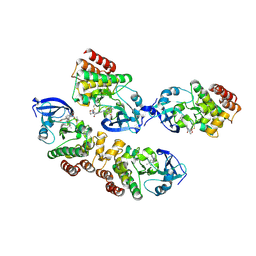 | |
4LL0
 
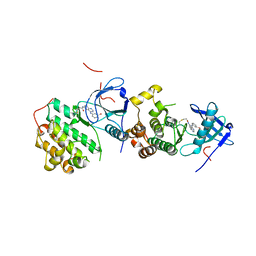 | | EGFR L858R/T790M in complex with PD168393 | | Descriptor: | Epidermal growth factor receptor, N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mechanism for activation of mutated epidermal growth factor receptors in lung cancer.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5CZH
 
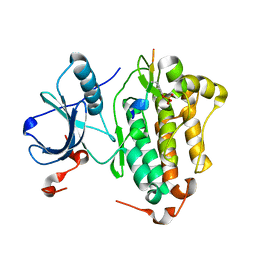 | |
5CZI
 
 | | EGFR L858R MUTANT IN COMPLEX WITH A SHC PEPTIDE SUBSTRATE | | Descriptor: | Epidermal growth factor receptor, SHC Peptide substrate | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | EGF-receptor specificity for phosphotyrosine-primed substrates provides signal integration with Src.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8IZP
 
 | | Multidrug resistance-associated protein 3 | | Descriptor: | ATP-binding cassette sub-family C member 3 | | Authors: | Yun, C.H, Gao, H.M. | | Deposit date: | 2023-04-07 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | structure of multidrug resistance-associated protein 3 at 3.31 Angstroms
To Be Published
|
|
8IZR
 
 | | Multidrug resistance-associated protein 3 | | Descriptor: | ATP-binding cassette sub-family C member 2 | | Authors: | Yun, C.H, Gao, H.M. | | Deposit date: | 2023-04-07 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | structure of multidrug resistance-associated protein 2 at 3.62 Angstroms resolution
To Be Published
|
|
5X26
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
1TGR
 
 | | Crystal Structure of mini-IGF-1(2) | | Descriptor: | Insulin-like growth factor IA | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2004-05-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42A crystal structure of mini-IGF-1(2): an analysis of the disulfide isomerization property and receptor binding property of IGF-1 based on the three-dimensional structure
Biochem.Biophys.Res.Commun., 326, 2004
|
|
1RFJ
 
 | | Crystal Structure of Potato Calmodulin PCM6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, calmodulin | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of potato calmodulin PCM6: the first report of the three-dimensional structure of a plant calmodulin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7YAZ
 
 | | Crystal structure of ZAK in complex with compound YH-186 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-[3-[(3~{S})-1-propanoylpyrrolidin-3-yl]oxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl]-1,2,3-triazol-1-yl]phenyl]-3-phenyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
7YAW
 
 | | Crystal structure of ZAK in complex with compound YH-180 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[3-[[5-[1-[2,6-bis(fluoranyl)-3-[(3-phenylphenyl)sulfonylamino]phenyl]-1,2,3-triazol-4-yl]-1~{H}-pyrazolo[3,4-b]pyridin-3-yl]oxy]propyl]propanamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
6LF6
 
 | | Crystal structure of ZmCGTa in complex with UDP | | Descriptor: | UDP-glycosyltransferase 708A6, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
