8SWK
 
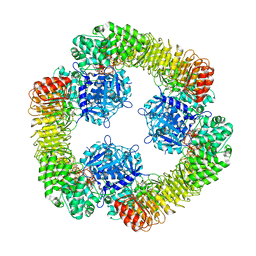 | | Cryo-EM structure of NLRP3 closed hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SXN
 
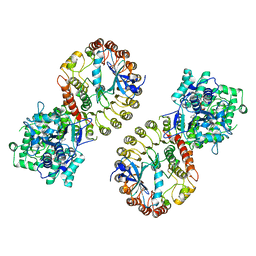 | | Structure of NLRP3 and NEK7 complex | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8BZM
 
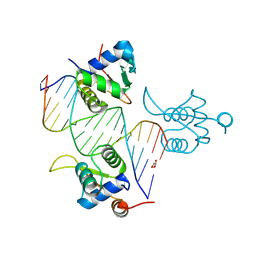 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
8UDL
 
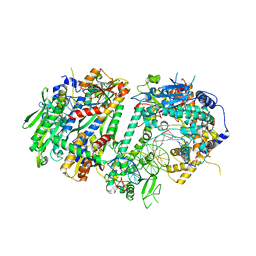 | | Human Mitochondrial DNA Polymerase Gamma Binary Complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*TP*AP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*T)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Park, J, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
6NJ0
 
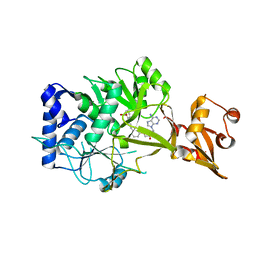 | | Wild-type E. coli MenE with bound m phenylether-linked analogue of OSB-AMS | | Descriptor: | 2-succinylbenzoate--CoA ligase, 5'-O-{3-[3-(2-carboxyphenyl)-3-oxopropyl]phenyl}adenosine | | Authors: | Si, Y, Yin, Y, French, J.B, Tonge, P.J. | | Deposit date: | 2019-01-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Non-Acyl Sulfamate Inhibitors of the Adenylate-Forming Enzyme MenE.
Biochemistry, 58, 2019
|
|
4ZTZ
 
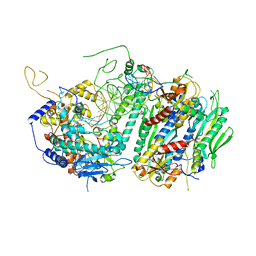 | |
4ZTU
 
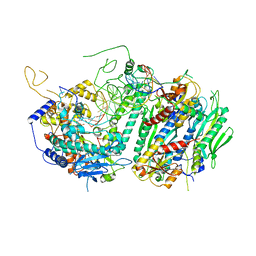 | | Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*AP*GP*AP*CP*GP*AP*GP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*GP*TP*AP*C)-3'), ... | | Authors: | Szymanski, M.R, Kuznestov, V.B, Shumate, C.K, Meng, Q, Lee, Y.-S, Patel, G, Patel, S.S, Yin, Y.W. | | Deposit date: | 2015-05-15 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for processivity and antiviral drug toxicity
in human mitochondrial DNA replicase
EMBO J., 34, 2015
|
|
2PI5
 
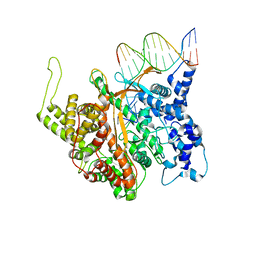 | | T7 RNA polymerase complexed with a phi10 promoter | | Descriptor: | 5'-D(*CP*TP*TP*C*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3', 5'-D(*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*T)-3', DNA-directed RNA polymerase | | Authors: | Kennedy, W.P, Momand, J.R, Yin, Y.W. | | Deposit date: | 2007-04-12 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism for de novo RNA synthesis and initiating nucleotide specificity by t7 RNA polymerase.
J.Mol.Biol., 370, 2007
|
|
2PI4
 
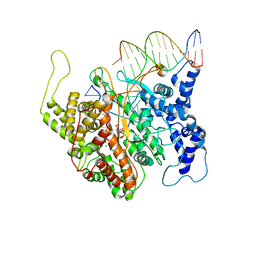 | | T7RNAP complexed with a phi10 protein and initiating GTPs. | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*CP*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3', 5'-D(*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*T)-3', ... | | Authors: | Kennedy, W.P, Momand, J.R, Yin, Y.W. | | Deposit date: | 2007-04-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for de novo RNA synthesis and initiating nucleotide specificity by t7 RNA polymerase.
J.Mol.Biol., 370, 2007
|
|
6DT0
 
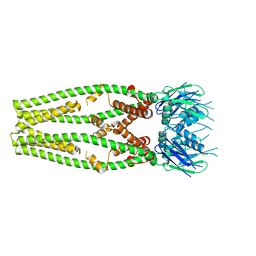 | | Cryo-EM structure of a mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Yoo, J, Wu, M, Yin, Y, Herzik, M.A.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a mitochondrial calcium uniporter.
Science, 361, 2018
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
7R6V
 
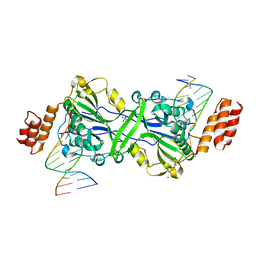 | | Human EXOG complexed with dRP-containing DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*CP*GP*TP*CP*AP*GP*AP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Szymanski, M.R, Yin, Y.W. | | Deposit date: | 2021-06-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Human EXOG possesses strong AP hydrolysis activity
To Be Published
|
|
3C8O
 
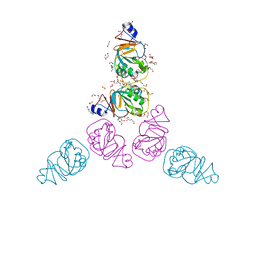 | | The Crystal Structure of RraA from PAO1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Regulator of ribonuclease activity A, ... | | Authors: | Luo, M, Niu, S, Yin, Y, Huang, A, Wang, D. | | Deposit date: | 2008-02-12 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of RraA from PAO1
To be published
|
|
8PE9
 
 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | Deposit date: | 2023-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
8KGG
 
 | | LPS-bound P2Y10 in complex with G13 | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Yin, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Insights into lysophosphatidylserine recognition and G alpha 12/13 -coupling specificity of P2Y10.
Cell Chem Biol, 2024
|
|
8PMF
 
 | |
8PMN
 
 | |
8PNA
 
 | |
8PM7
 
 | |
8PNC
 
 | |
8PMC
 
 | |
8PMV
 
 | |
1PES
 
 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
8UDK
 
 | | Human Mitochondrial DNA Polymerase gamma R853A Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (24-MER), DNA (28-MER), ... | | Authors: | Park, J, Herrmann, G.K, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
