6P4Q
 
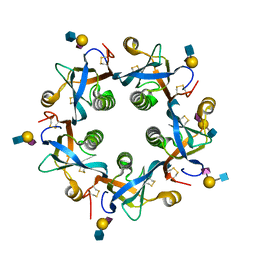 | | Salmonella typhi PltB Homopentamer N29K Mutant with Neu5Ac-alpha-2-3-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
6P4N
 
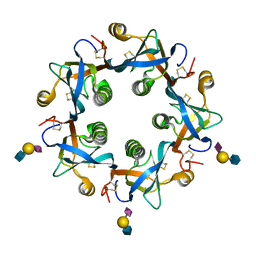 | | Salmonella typhi PltB Homopentamer with Neu5Ac-alpha-2-6-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
6P4S
 
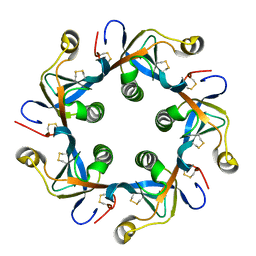 | | Salmonella typhi PltB Homopentamer T65I Mutant | | Descriptor: | Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
6P4P
 
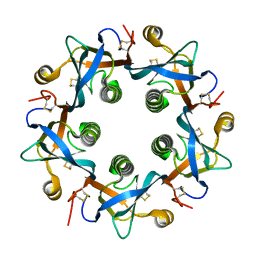 | | Salmonella typhi PltB Homopentamer N29K Mutant | | Descriptor: | Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
8JPY
 
 | |
8SG2
 
 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
6X4X
 
 | | B24Y DKP insulin | | Descriptor: | Insulin, Insulin chain A | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2020-05-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evolution of insulin at the edge of foldability and its medical implications.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
1G47
 
 | | 1ST LIM DOMAIN OF PINCH PROTEIN | | Descriptor: | PINCH PROTEIN, ZINC ION | | Authors: | Velyvis, A, Yang, Y, Wu, C, Qin, J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain.
J.Biol.Chem., 276, 2001
|
|
2EEM
 
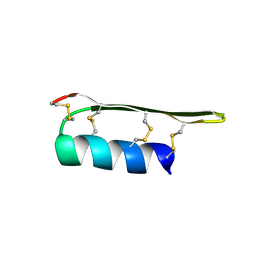 | |
6UPG
 
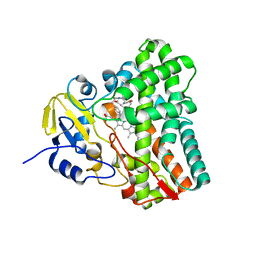 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with cYF-4-OMe | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(4-methoxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nguyen, R.C.D, Yang, Y, Liu, A. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Substrate-Assisted Hydroxylation and O-Demethylation in the Peroxidase-like Cytochrome P450 Enzyme CYP121
Acs Catalysis, 10, 2020
|
|
6UPI
 
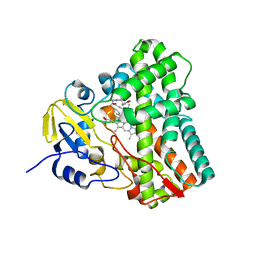 | | Crystal structure of Mycobacterium tuberculosis CYP121 bound with a hydroxylated intermediate of cYF-4-OMe | | Descriptor: | (3S,6S)-3-{[4-(hydroxymethoxy)phenyl]methyl}-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nguyen, R.C.D, Yang, Y, Liu, A. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Substrate-Assisted Hydroxylation and O-Demethylation in the Peroxidase-like Cytochrome P450 Enzyme CYP121
Acs Catalysis, 10, 2020
|
|
1MR0
 
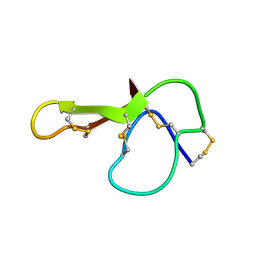 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
7N0D
 
 | |
5C56
 
 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
6XH7
 
 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
5JWA
 
 | | the structure of malaria PfNDH2 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y, Yang, Y.Q, Li, X.L, Yu, J, Ge, J.P, Li, J, Rao, Y, Yang, M.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
7YW7
 
 | |
7YW8
 
 | |
1JSG
 
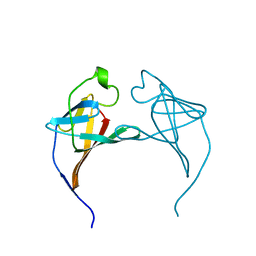 | | CRYSTAL STRUCTURE OF P14TCL1, AN ONCOGENE PRODUCT INVOLVED IN T-CELL PROLYMPHOCYTIC LEUKEMIA, REVEALS A NOVEL B-BARREL TOPOLOGY | | Descriptor: | ONCOGENE PRODUCT P14TCL1 | | Authors: | Hoh, F, Yang, Y.-S, Guignard, L, Padilla, A, Stern, R.-H, Lhoste, J.-M, Van Tilbeurgh, H. | | Deposit date: | 1997-12-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of p14TCL1, an oncogene product involved in T-cell prolymphocytic leukemia, reveals a novel beta-barrel topology.
Structure, 6, 1998
|
|
5KWQ
 
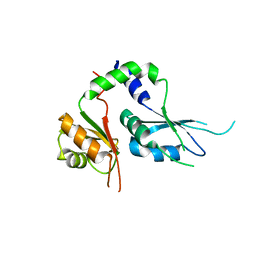 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
3IXT
 
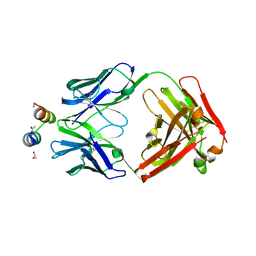 | | Crystal Structure of Motavizumab Fab Bound to Peptide Epitope | | Descriptor: | 1,2-ETHANEDIOL, Fusion glycoprotein F1, Motavizumab Fab heavy chain, ... | | Authors: | McLellan, J.S, Chen, M, Kim, A, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2009-09-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of respiratory syncytial virus neutralization by motavizumab.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1UXD
 
 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1T0P
 
 | | Structural Basis of ICAM recognition by integrin alpahLbeta2 revealed in the complex structure of binding domains of ICAM-3 and alphaLbeta2 at 1.65 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, Intercellular adhesion molecule-3, ... | | Authors: | Song, G, Yang, Y.T, Liu, J.H, Shimaoko, M, Springer, T.A, Wang, J.H. | | Deposit date: | 2004-04-12 | | Release date: | 2005-03-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alphaLbeta2.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
