7MEV
 
 | | Human Apex/Ref1 monomer with C138A mutation | | Descriptor: | DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial, GLYCEROL, ... | | Authors: | Nam, Y.W, Yang, S. | | Deposit date: | 2021-04-07 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Development of Novel Apurinic/Aprymidinic Endonuclease/Redox-factor 1 Inhibitors for the Treatment of Human Melanoma
To Be Published
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5KZF
 
 | |
8T6V
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T6U
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GUD
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUB
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUA
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E542K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ZM6
 
 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
4RL4
 
 | |
8YKI
 
 | | FGFR-1 in complex with ligand tasurgratinib | | Descriptor: | CHLORIDE ION, Fibroblast growth factor receptor 1, Tasurgratinib | | Authors: | Ikemori-Kawada, M, Watanabe Miyano, S. | | Deposit date: | 2024-03-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Antitumor Activity of Tasurgratinib as an Orally Available FGFR1-3 Inhibitor in Cholangiocarcinoma Models With FGFR2-fusion.
Anticancer Res., 44, 2024
|
|
8QFU
 
 | | Diels-Alderase AbyU mutant - Y76F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Tiwari, K, Burton, N.M, Yang, S, Race, P.R. | | Deposit date: | 2023-09-05 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Diels-Alderase AbyU mutant Y76F at 1.56 Angstroms resolution.
To Be Published
|
|
6UZT
 
 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
5JLE
 
 | | Crystal structure of SETD2 bound to SAH | | Descriptor: | Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, H, Yang, S, Zheng, X. | | Deposit date: | 2016-04-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
1N03
 
 | | Model for Active RecA Filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RecA protein | | Authors: | VanLoock, M.S, Yu, X, Yang, S, Lai, A.L, Low, C, Campbell, M.J, Egelman, E.H. | | Deposit date: | 2002-10-10 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | ATP-Mediated Conformational Changes in the RecA Filament
Structure, 11, 2003
|
|
5JLB
 
 | |
3JBM
 
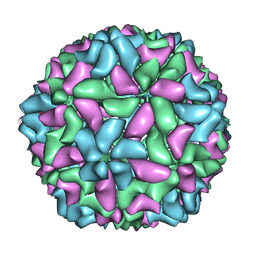 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
3LLP
 
 | | 1.8 Angstrom human fascin 1 crystal structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, Fascin, ... | | Authors: | Chen, L, Yang, S, Jakoncic, J, Zhang, J.J, Huang, X.-Y. | | Deposit date: | 2010-01-29 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Migrastatin analogues target fascin to block tumour metastasis.
Nature, 464, 2010
|
|
2X79
 
 | | Inward facing conformation of Mhp1 | | Descriptor: | Hydantoin permease | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
3RZE
 
 | | Structure of the human histamine H1 receptor in complex with doxepin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3E)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, (3Z)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, ... | | Authors: | Shimamura, T, Han, G.W, Shiroishi, M, Weyand, S, Tsujimoto, H, Winter, G, Katritch, V, Abagyan, R, Cherezov, V, Liu, W, Kobayashi, T, Stevens, R, Iwata, S, GPCR Network (GPCR) | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the human histamine H1 receptor complex with doxepin.
Nature, 475, 2011
|
|
1LQV
 
 | | Crystal structure of the Endothelial protein C receptor with phospholipid in the groove in complex with Gla domain of protein C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-05-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
