7RE5
 
 | |
6KI0
 
 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
1ALB
 
 | |
6V97
 
 | | Kindlin-3 double deletion mutant short form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
6V9G
 
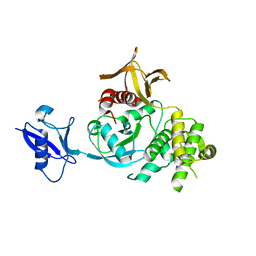 | | Kindlin-3 double deletion mutant long form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
1AON
 
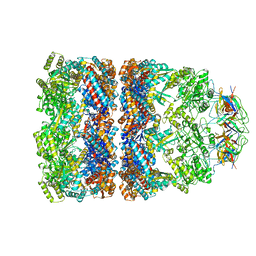 | | CRYSTAL STRUCTURE OF THE ASYMMETRIC CHAPERONIN COMPLEX GROEL/GROES/(ADP)7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GROEL, GROEL/GROES COMPLEX, ... | | Authors: | Xu, Z, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1997-07-08 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex.
Nature, 388, 1997
|
|
7SPD
 
 | |
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
5GMZ
 
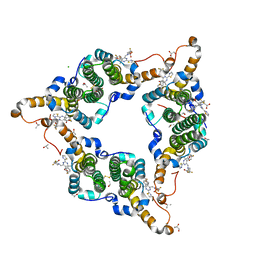 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
5HMN
 
 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
3FGA
 
 | | Structural Basis of PP2A and Sgo interaction | | Descriptor: | MANGANESE (II) ION, MICROCYSTIN-LR, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Xu, Z, Xu, W. | | Deposit date: | 2008-12-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the PP2A-shugoshin interaction
Mol.Cell, 35, 2009
|
|
1FX3
 
 | |
4HPH
 
 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOW
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOX
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 in complex with Tris
to be published
|
|
4HOZ
 
 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HP5
 
 | | The crystal structure of isomaltulose synthase mutant E295A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of isomaltulose synthase mutant E295A from Erwinia rhapontici NX5 in complex with D-glucose
to be published
|
|
5F47
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with trehalose | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
4RE1
 
 | | Crystal structure of human TEAD1 and disulfide-engineered YAP | | Descriptor: | CHLORIDE ION, Transcriptional enhancer factor TEF-1, Yorkie homolog | | Authors: | Xu, Z, Zhou, Z. | | Deposit date: | 2014-09-21 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides.
Faseb J., 29, 2015
|
|
5U08
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
2F42
 
 | |
1NFG
 
 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
5F46
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample, apoenzyme form | | Descriptor: | CHLORIDE ION, aminoglycoside acetyltransferase meta-AAC0020 | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
