8IUK
 
 | | Cryo-EM structure of the PGF2-alpha-bound human PTGFR-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8H0P
 
 | | Structure of the NMB30-NMBR and Gq complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Xu, Y, Liu, H, Cai, H, Xu, H.E, Yin, W. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular recognition of itch-associated neuropeptides by bombesin receptors
Cell Res., 33, 2023
|
|
8H0Q
 
 | | Structure of the GRP14-27-GRPR-Gq complex | | Descriptor: | CHOLESTEROL, G-alpha q, GRP, ... | | Authors: | Li, C, Xu, Y, Liu, H, Cai, H, Xu, H.E, Yin, W. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of itch-associated neuropeptides by bombesin receptors
Cell Res., 33, 2023
|
|
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
8WTF
 
 | | The Crystal Structure of IRAK4 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of IRAK4 from Biortus
To Be Published
|
|
8X87
 
 | |
8X88
 
 | |
8XPV
 
 | | The Crystal Structure of EphA2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, Ephrin type-A receptor 2, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-01-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of EphA2 from Biortus.
To Be Published
|
|
8Y33
 
 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
8YHW
 
 | | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, MAGNESIUM ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus
To Be Published
|
|
8YHF
 
 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
8YHL
 
 | | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, ACETATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus
To Be Published
|
|
8YGY
 
 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
2I7U
 
 | | Structural and Dynamical Analysis of a Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets | | Descriptor: | Four-alpha-helix bundle | | Authors: | Ma, D, Brandon, N.R, Cui, T, Bondarenko, V, Canlas, C, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part I: structural and dynamical analyses.
Biophys.J., 94, 2008
|
|
5YTK
 
 | | Crystal structure of SIRT3 bound to a leucylated AceCS2 | | Descriptor: | AceCS2-KLeu, LEUCINE, LYSINE, ... | | Authors: | Li, J, Gong, W, Xu, Y. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations
Cell Metab., 27, 2018
|
|
8JPA
 
 | |
8J98
 
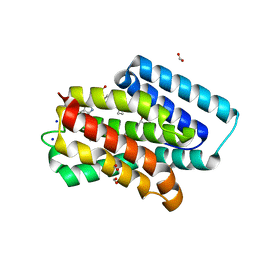 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo backbone design for monomerization of near-infrared fluorescent protein
To Be Published
|
|
3UAN
 
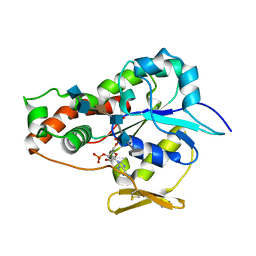 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8QMZ
 
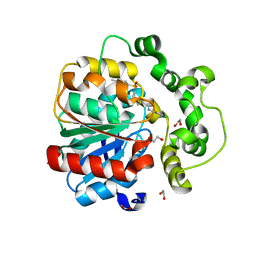 | | Soluble epoxide hydrolase in complex with RK4 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-2-(4-methylphenyl)sulfonyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-5-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
8QN0
 
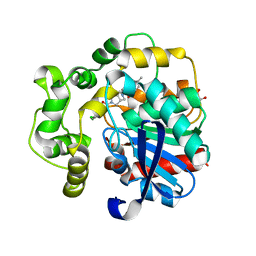 | | Soluble epoxide hydrolase in complex with RK3 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-5-(4-methylphenyl)sulfonyl-1,3,3~{a},4,6,6~{a}-hexahydropyrrolo[3,4-c]pyrrole-2-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
3EK9
 
 | | SPRY Domain-containing SOCS Box Protein 2: Crystal Structure and Residues Critical for Protein Binding | | Descriptor: | GLYCEROL, SPRY domain-containing SOCS box protein 2 | | Authors: | Kuang, Z, Yao, S, Xu, Y, Garrett, T.J.P, Norton, R.S. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SPRY domain-containing SOCS box protein 2: crystal structure and residues critical for protein binding.
J.Mol.Biol., 386, 2009
|
|
3FPP
 
 | | Crystal structure of E.coli MacA | | Descriptor: | Macrolide-specific efflux protein macA | | Authors: | Yum, S, Xu, Y, Piao, S, Ha, N.-C. | | Deposit date: | 2009-01-06 | | Release date: | 2009-04-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump
J.Mol.Biol., 387, 2009
|
|
3G5V
 
 | | Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR | | Descriptor: | 806 light chain, 808 heavy chain, ACETATE ION, ... | | Authors: | Garrett, T.P.J, Burgess, A.W, Huyton, T, Xu, Y. | | Deposit date: | 2009-02-05 | | Release date: | 2010-02-09 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Antibodies specifically targeting a locally misfolded region of tumor associated EGFR
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
