1U4A
 
 | | Solution structure of human SUMO-3 C47S | | Descriptor: | Ubiquitin-like protein SMT3A | | Authors: | Ding, H, Xu, Y, Dai, H, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2004-07-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human SUMO-3 C47S and Its Binding Surface for Ubc9
Biochemistry, 44, 2005
|
|
2XI4
 
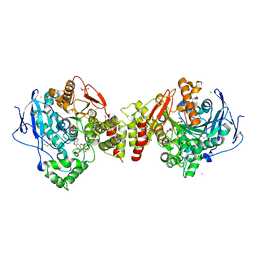 | | Torpedo californica Acetylcholinesterase in Complex with Aflatoxin B1 (Orthorhombic Space Group) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, AFLATOXIN B1, ... | | Authors: | Sanson, B, Colletier, J.P, Xu, Y, Lang, P.T, Jiang, H, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2010-06-28 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Backdoor Opening Mechanism in Acetylcholinesterase Based on X-Ray Crystallography and Molecular Dynamics Simulations.
Protein Sci., 20, 2011
|
|
4DBB
 
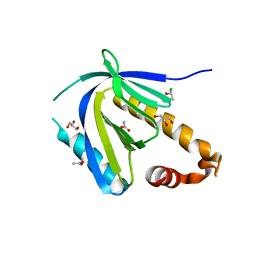 | | The PTB domain of Mint1 is autoinhibited by a helix in the C-terminal linker region | | Descriptor: | ACETIC ACID, Amyloid beta A4 precursor protein-binding family A member 1, CHLORIDE ION, ... | | Authors: | Tomchick, D.R, Rizo, J, Ho, A, Xu, Y. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Autoinhibition of Mint1 adaptor protein regulates amyloid precursor protein binding and processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5SXU
 
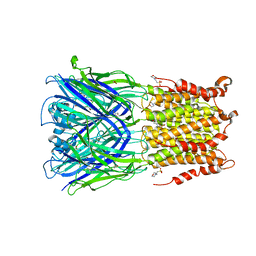 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a desensitized state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, 3-AMINOPROPANE, ... | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
5SXV
 
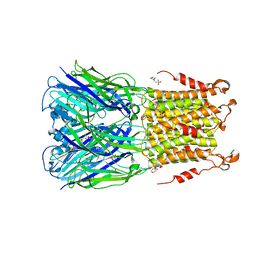 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a resting state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, Cys-loop ligand-gated ion channel | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
4DUG
 
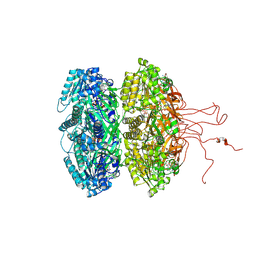 | | Crystal Structure of Circadian Clock Protein KaiC E318A Mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION, ... | | Authors: | Egli, M, Mori, T, Pattanayek, R, Xu, Y, Qin, X, Johnson, C.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Dephosphorylation of the Core Clock Protein KaiC in the Cyanobacterial KaiABC Circadian Oscillator Proceeds via an ATP Synthase Mechanism.
Biochemistry, 51, 2012
|
|
5TIH
 
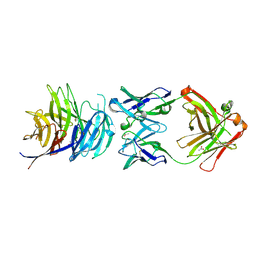 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | ACETATE ION, CyRPA antibody Fab Heavy Chain, CyRPA antibody Fab Light Chain, ... | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5TIK
 
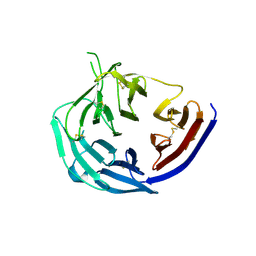 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | Cysteine-rich protective antigen | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5U8Q
 
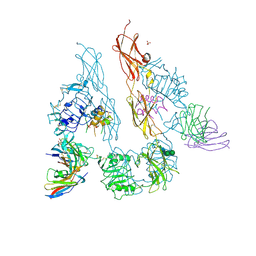 | |
5U8R
 
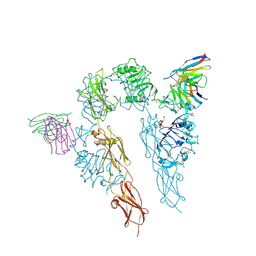 | |
5C56
 
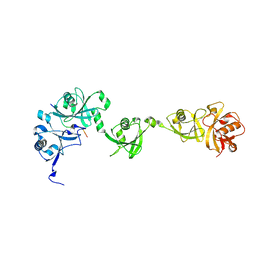 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
4PY4
 
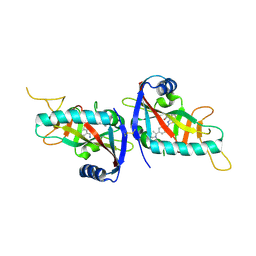 | |
4F8H
 
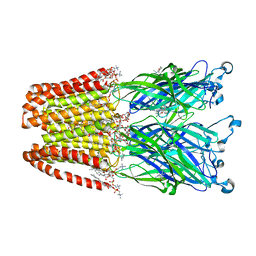 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | Descriptor: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | Authors: | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
5DX4
 
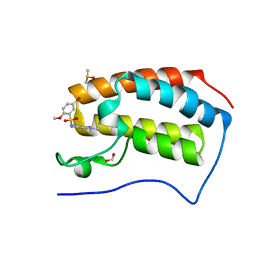 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-2-methoxybenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
7Y8R
 
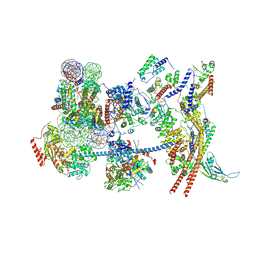 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
3U0J
 
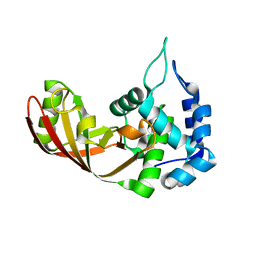 | |
3U9G
 
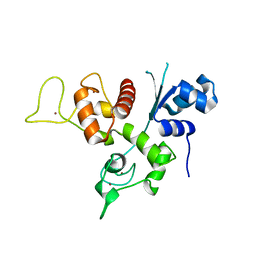 | | Crystal structure of the Zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1QCQ
 
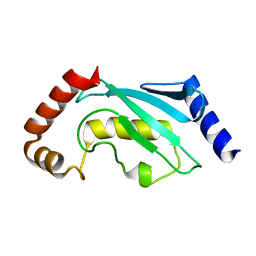 | | UBIQUITIN CONJUGATING ENZYME | | Descriptor: | PROTEIN (UBIQUITIN CONJUGATING ENZYME) | | Authors: | Cook, W.J, Jeffrey, L.C, Xu, Y, Chau, V. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4.
Biochemistry, 32, 1993
|
|
1R19
 
 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (Apo structure) | | Descriptor: | fibrinogen-binding protein SdrG | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
1RTK
 
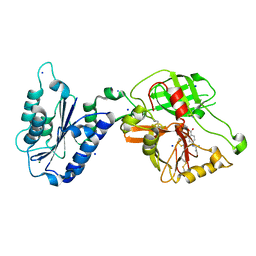 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, Complement factor B Bb fragment, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1R17
 
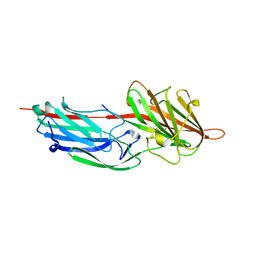 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | | Descriptor: | CALCIUM ION, fibrinogen-binding protein SdrG, fibrinopeptide B | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
1RRK
 
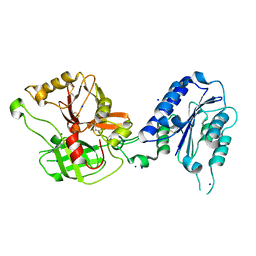 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RS0
 
 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with Di-isopropyl-phosphate (DIP) | | Descriptor: | Complement factor B, DIISOPROPYL PHOSPHONATE, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
3WLF
 
 | | Crystal structure of (R)-carbonyl reductase from Candida Parapsilosis in complex with (R)-1-phenyl-1,2-ethanediol | | Descriptor: | (1R)-1-phenylethane-1,2-diol, (R)-specific carbonyl reductase, ZINC ION | | Authors: | Wang, S.S, Nie, Y, Xu, Y, Zhang, R.Z, Huang, C.H, Chan, H.C, Guo, R.T, Xiao, R. | | Deposit date: | 2013-11-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unconserved substrate-binding sites direct the stereoselectivity of medium-chain alcohol dehydrogenase
Chem.Commun.(Camb.), 50, 2014
|
|
3WLE
 
 | | Crystal structure of (R)-carbonyl reductase from Candida Parapsilosis in complex with NAD | | Descriptor: | (R)-specific carbonyl reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Wang, S.S, Nie, Y, Xu, Y, Zhang, R.Z, Huang, C.H, Chan, H.C, Guo, R.T, Xiao, R. | | Deposit date: | 2013-11-08 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Unconserved substrate-binding sites direct the stereoselectivity of medium-chain alcohol dehydrogenase
Chem.Commun.(Camb.), 50, 2014
|
|
