6XNE
 
 | |
6XNM
 
 | |
6XNF
 
 | |
6XNL
 
 | |
4DBB
 
 | | The PTB domain of Mint1 is autoinhibited by a helix in the C-terminal linker region | | Descriptor: | ACETIC ACID, Amyloid beta A4 precursor protein-binding family A member 1, CHLORIDE ION, ... | | Authors: | Tomchick, D.R, Rizo, J, Ho, A, Xu, Y. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Autoinhibition of Mint1 adaptor protein regulates amyloid precursor protein binding and processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1LRZ
 
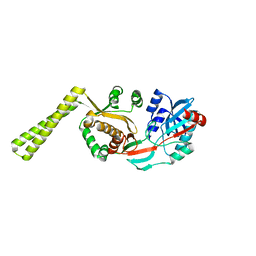 | | x-ray crystal structure of staphylococcus aureus femA | | Descriptor: | factor essential for expression of methicillin resistance | | Authors: | Benson, T, Prince, D, Mutchler, V, Curry, K, Ho, A, Sarver, R, Hagadorn, J, Choi, G, Garlick, R. | | Deposit date: | 2002-05-16 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of Staphylococcus aureus FemA.
Structure, 10, 2002
|
|
1TKN
 
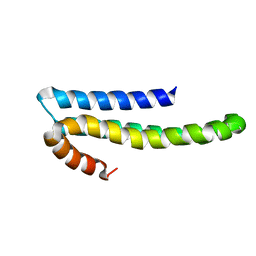 | | Solution structure of CAPPD*, an independently folded extracellular domain of human Amyloid-beta Precursor Protein | | Descriptor: | Amyloid beta A4 protein | | Authors: | Dulubova, I, Ho, A, Huryeva, I, Sudhof, T.C, Rizo, J. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of an independently folded extracellular domain of human amyloid-beta precursor protein.
Biochemistry, 43, 2004
|
|
