3CVF
 
 | | Crystal Structure of the carboxy terminus of Homer3 | | Descriptor: | Homer protein homolog 3 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
3CVE
 
 | | Crystal Structure of the carboxy terminus of Homer1 | | Descriptor: | Homer protein homolog 1 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
3DY6
 
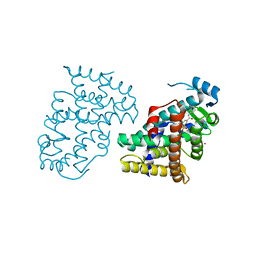 | | PPARdelta complexed with an anthranilic acid partial agonist | | Descriptor: | 2-({[3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)phenyl]carbonyl}amino)benzoic acid, IODIDE ION, Peroxisome proliferator-activated receptor delta | | Authors: | Lambert, M.L, Xu, R, Shearer, B.G, Wilson, T.M. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a novel class of PPARdelta partial agonists
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8W4A
 
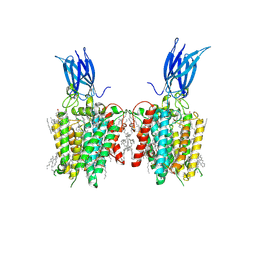 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-08-23 | | Release date: | 2024-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4KUL
 
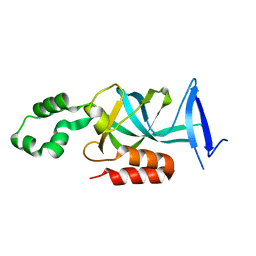 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5UOY
 
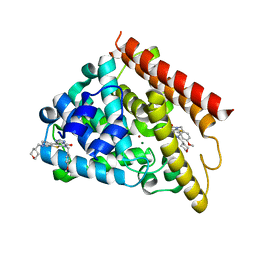 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 16j (6-(4-Methoxybenzyl)-9-((tetrahydro-2H-pyran-4-yl)methyl)-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-methoxyphenyl)methyl]-9-[(oxan-4-yl)methyl]-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
3G0B
 
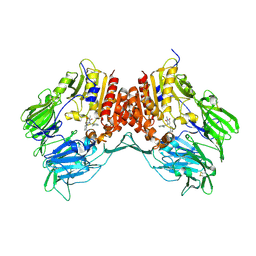 | | Crystal structure of dipeptidyl peptidase IV in complex with TAK-322 | | Descriptor: | 2-({6-[(3R)-3-aminopiperidin-1-yl]-3-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl}methyl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
3G0C
 
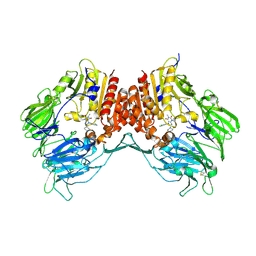 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(2-chlorobenzyl)-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-1H-purine-2,6-dione, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
5UP0
 
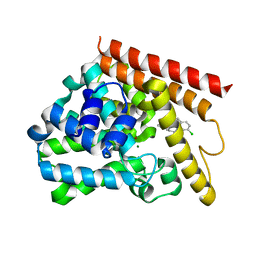 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 3 (6-(4-chlorobenzyl)-8,9,10,11-tetrahydrobenzo[4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-chlorophenyl)methyl]-8,9,10,11-tetrahydro[1]benzothieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
5EX7
 
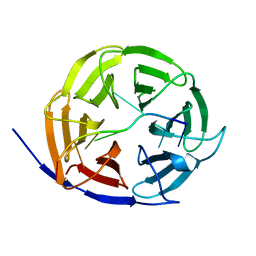 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
5H1K
 
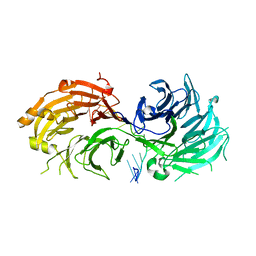 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | Descriptor: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | Authors: | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
1NW3
 
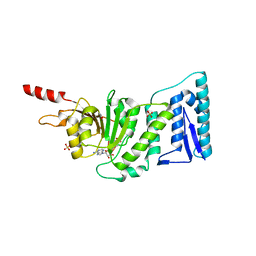 | | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Min, J.R, Feng, Q, Li, Z.H, Zhang, Y, Xu, R.M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase
Cell(Cambridge,Mass.), 112, 2003
|
|
1URK
 
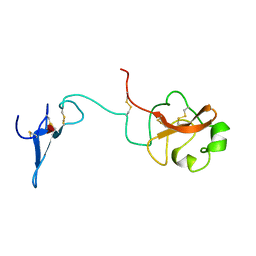 | | SOLUTION STRUCTURE OF THE AMINO TERMINAL FRAGMENT OF UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | PLASMINOGEN ACTIVATOR, alpha-L-fucopyranose | | Authors: | Hansen, A.P, Petros, A.M, Meadows, R.P, Nettesheim, D.G, Mazar, A.P, Olejniczak, E.T, Xu, R.X, Pederson, T.M, Henkin, J, Fesik, S.W. | | Deposit date: | 1994-01-10 | | Release date: | 1995-05-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator.
Biochemistry, 33, 1994
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2ONC
 
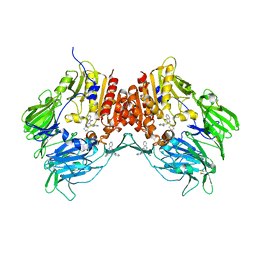 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
1OO0
 
 | |
3OOI
 
 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1YMT
 
 | | Mouse SF-1 LBD | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, Nuclear receptor 0B2, Steroidogenic factor 1 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, Juzumiene, D, Bynum, J.M, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
3QKL
 
 | | Spirochromane Akt Inhibitors | | Descriptor: | GSK-3 beta peptide, N-{(2S)-3-[(3S)-8',9'-dihydro-1H,3'H-spiro[piperidine-3,7'-pyrano[3,2-e]indazol]-1-yl]-2-hydroxypropyl}-N-(2-ethoxyethyl)-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PLZ
 
 | | Human LRH1 LBD bound to GR470 | | Descriptor: | (3aS,6aR)-5-[(4E)-oct-4-en-4-yl]-N,4-diphenyl-2,3,6,6a-tetrahydropentalen-3a(1H)-amine, 1,2-ETHANEDIOL, FTZ-F1 related protein, ... | | Authors: | Williams, S.P, Xu, R, Zuercher, W.J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Agonists of the Orphan Nuclear Receptors Steroidogenic Factor-1 (SF-1, NR5A1) and Liver Receptor Homologue-1 (LRH-1, NR5A2).
J.Med.Chem., 54, 2011
|
|
3QKK
 
 | | Spirochromane Akt Inhibitors | | Descriptor: | Glycogen synthase kinase-3 beta, N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(2R)-6-hydroxy-4-oxo-3,4-dihydro-1'H-spiro[chromene-2,3'-piperidin]-1'-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5C3I
 
 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | Descriptor: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | Authors: | Wang, H, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
3NTI
 
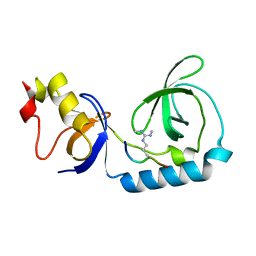 | | Crystal structure of Tudor and Aubergine [R15(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
3NTK
 
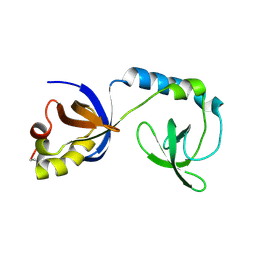 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
