7XOC
 
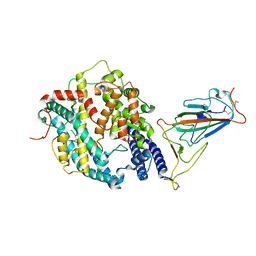 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO9
 
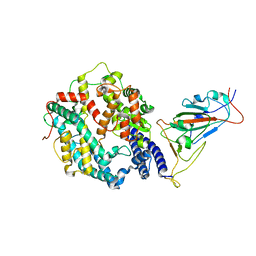 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XW5
 
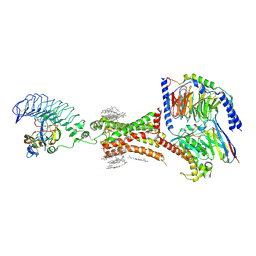 | | TSHR-thyroid stimulating hormone-Gs-ML109 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Glycoprotein hormones alpha chain, ... | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
1ZH7
 
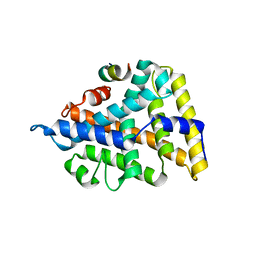 | | Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP | | Descriptor: | Orphan nuclear receptor NR5A2, nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Suino, K, Kovach, A, Daugherty, J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2005-04-22 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6C9G
 
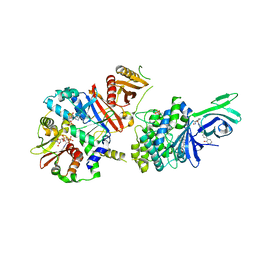 | | AMP-activated protein kinase bound to pharmacological activator R739 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9F
 
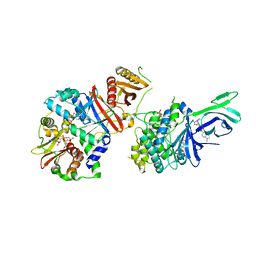 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9H
 
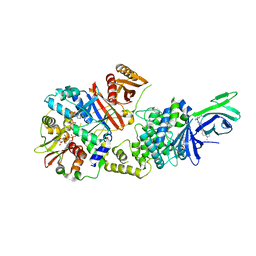 | | non-phosphorylated AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
7FIG
 
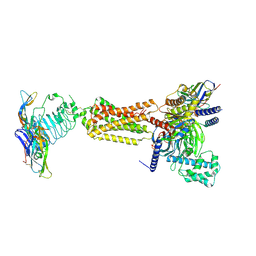 | | luteinizing hormone/choriogonadotropin receptor(S277I)-chorionic gonadotropin-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Camelid antibody VHH fragment Nb35, Choriogonadotropin subunit beta 3, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7JHG
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JHH
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
4ME2
 
 | | Crystal Structure of THA8 protein from Brachypodium distachyon | | Descriptor: | Uncharacterized protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Melcher, K, Xu, H.E. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3C4M
 
 | |
3N93
 
 | | Crystal structure of human CRFR2 alpha extracellular domain in complex with Urocortin 3 | | Descriptor: | GLYCEROL, Maltose binding protein-CRFR2 alpha, Urocortin-3, ... | | Authors: | Pal, K, Swaminathan, K, Pioszak, A.A, Xu, H.E. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of ligand selectivity in human CRFR1 and CRFR2 alpha extracellular domain
To be Published
|
|
4LRH
 
 | | Crystal structure of human folate receptor alpha in complex with folic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, Folate receptor alpha | | Authors: | Ke, J, Chen, C, Zhou, X.E, Yi, W, Brunzelle, J.S, Li, J, Young, E.-L, Xu, H.E, Melcher, K. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition of folic acid by folate receptors.
Nature, 500, 2013
|
|
2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
4LOG
 
 | | The crystal structure of the orphan nuclear receptor PNR ligand binding domain fused with MBP | | Descriptor: | Maltose ABC transporter periplasmic protein and NR2E3 protein chimeric construct | | Authors: | Tan, M.E, Zhou, X.E, Soon, F.-F, Li, X, Li, J, Yong, E.-L, Melcher, K, Xu, H.E. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Orphan Nuclear Receptor NR2E3/PNR Ligand Binding Domain Reveals a Dimeric Auto-Repressed Conformation.
Plos One, 8, 2013
|
|
3HND
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP and substrate GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3HNF
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effectors TTP and dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3BZU
 
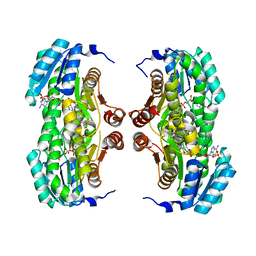 | | Crystal structure of human 11-beta-hydroxysteroid dehydrogenase(HSD1) in complex with NADP and thiazolone inhibitor | | Descriptor: | (5S)-2-{[(1S)-1-(2-fluorophenyl)ethyl]amino}-5-methyl-5-(trifluoromethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Min, X, Sudom, A, Xu, H, Wang, Z, Walker, N.P. | | Deposit date: | 2008-01-18 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization and pharmacodynamic effects of an orally active 11beta-hydroxysteroid dehydrogenase type 1 inhibitor.
Chem.Biol.Drug Des., 71, 2008
|
|
3HNC
 
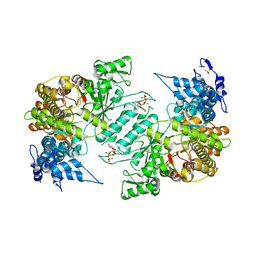 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP | | Descriptor: | MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, SULFATE ION, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3HNE
 
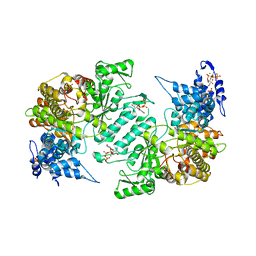 | | Crystal structure of human ribonucleotide reductase 1 bound to the effectors TTP and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4YZ6
 
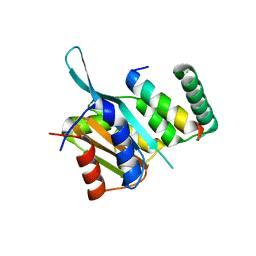 | | Crystal Structure of Myc3[44-238] from Arabidopsis in complex with Jaz1 peptide [200-221] | | Descriptor: | Protein TIFY 10A, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J, Xu, H.E, Melcher, K, HE, S.Y. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4N2Q
 
 | | Crystal structure of THA8 in complex with Zm4 RNA | | Descriptor: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4N2S
 
 | | Crystal Structure of THA8 in complex with Zm1a-6 RNA | | Descriptor: | THA8 RNA binding protein, Zm1a-6 RNA | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2PL5
 
 | | Crystal Structure of Homoserine O-acetyltransferase from Leptospira interrogans | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Liu, L, Wang, M, Wang, Y, Wei, Z, Xu, H, Gong, W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans
Biochem.Biophys.Res.Commun., 363, 2007
|
|
