7XJY
 
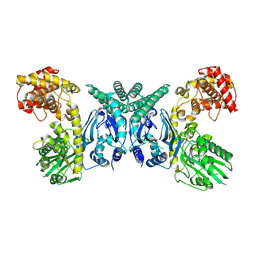 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase (apo form) | | 分子名称: | Glycine--tRNA ligase | | 著者 | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | 登録日 | 2022-04-19 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK1
 
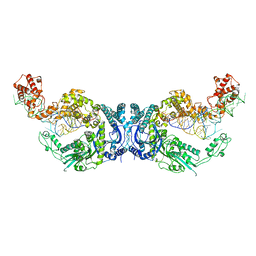 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | | 分子名称: | Glycine--tRNA ligase, tRNA(gly) | | 著者 | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | 登録日 | 2022-04-19 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK0
 
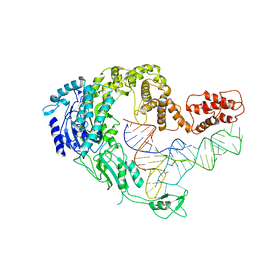 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA locked state) | | 分子名称: | Glycine--tRNA ligase, tRNA(gly) | | 著者 | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | 登録日 | 2022-04-19 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-13 | | 公開日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
5B7W
 
 | |
6M0W
 
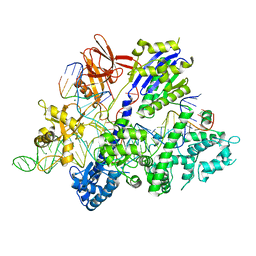 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | 分子名称: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0X
 
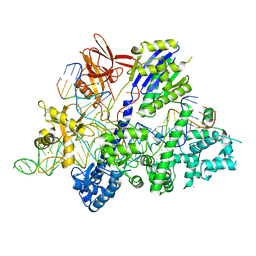 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.561 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
1K6U
 
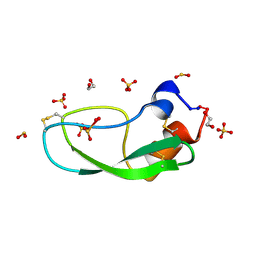 | | Crystal Structure of Cyclic Bovine Pancreatic Trypsin Inhibitor | | 分子名称: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | 著者 | Botos, I, Wu, Z, Lu, W, Wlodawer, A. | | 登録日 | 2001-10-17 | | 公開日 | 2001-12-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal structure of a cyclic form of bovine pancreatic trypsin inhibitor.
FEBS Lett., 509, 2001
|
|
5VTO
 
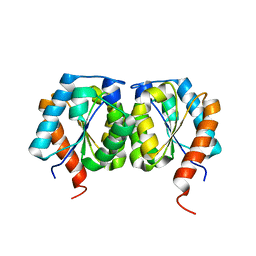 | |
1N8X
 
 | | Solution structure of HIV-1 Stem Loop SL1 | | 分子名称: | HIV-1 STEM LOOP SL1 MONOMERIC RNA | | 著者 | Lawrence, D.C, Stover, C.C, Noznitsky, J, Wu, Z, Summers, M.F. | | 登録日 | 2002-11-21 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Intact Stem and Bulge of HIV-1 Psi-RNA Stem-Loop SL1
J.Mol.Biol., 326, 2003
|
|
7X63
 
 | | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex | | 分子名称: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | 著者 | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | 登録日 | 2022-03-06 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7X66
 
 | | SARS-CoV-2-Omicron-RBD and BD-236-GWP/P-VK antibody complex | | 分子名称: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | 著者 | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | 登録日 | 2022-03-06 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | SARS-CoV-2-Omicron-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7XIL
 
 | | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B38 Fab heavy chain, B38 Fab light chain, ... | | 著者 | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | 登録日 | 2022-04-13 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
7XIK
 
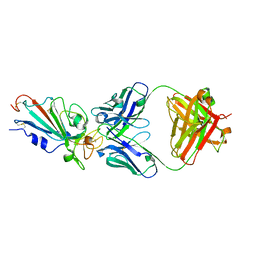 | | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex | | 分子名称: | B38 Fab heavy chain, B38 Fab light chain, Spike protein S1 | | 著者 | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | 登録日 | 2022-04-13 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
6M0V
 
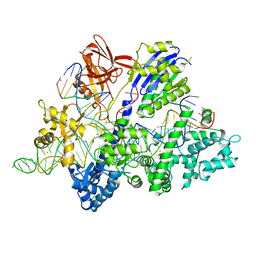 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-22 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
3IR1
 
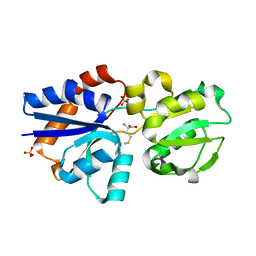 | | Crystal Structure of Lipoprotein GNA1946 from Neisseria meningitidis | | 分子名称: | METHIONINE, Outer membrane lipoprotein GNA1946, SULFATE ION | | 著者 | Yang, X, Wu, Z, Wang, X, Shen, Y. | | 登録日 | 2009-08-21 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of lipoprotein GNA1946 from Neisseria meningitidis
J.Struct.Biol., 168, 2009
|
|
7CUJ
 
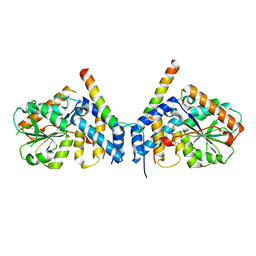 | | Crystal structure of fission yeast Ccq1 and Tpz1 | | 分子名称: | Coiled-coil quantitatively-enriched protein 1, Protection of telomeres protein tpz1 | | 著者 | Sun, H, Wu, Z, Wu, J, Lei, M. | | 登録日 | 2020-08-23 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUH
 
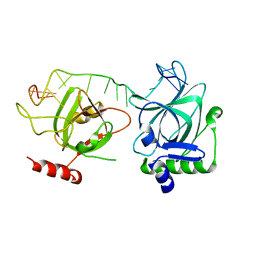 | | Crystal structure of fission yeast Pot1 and ssDNA | | 分子名称: | Protection of telomeres protein 1, Telomere single-strand DNA | | 著者 | Sun, H, Wu, Z, Wu, J, Lei, M. | | 登録日 | 2020-08-23 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUI
 
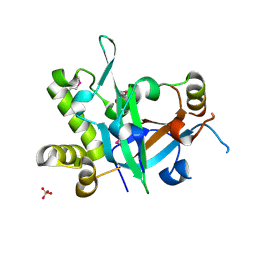 | | Crystal structure of fission yeast Pot1 and Tpz1 | | 分子名称: | Protection of telomeres protein 1, Protection of telomeres protein tpz1, SULFATE ION | | 著者 | Sun, H, Wu, Z, Wu, J, Lei, M. | | 登録日 | 2020-08-23 | | 公開日 | 2021-08-25 | | 最終更新日 | 2022-09-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
3R0F
 
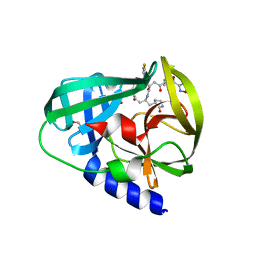 | | Human enterovirus 71 3C protease mutant H133G in complex with rupintrivir | | 分子名称: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | 登録日 | 2011-03-08 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.3083 Å) | | 主引用文献 | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
3QZR
 
 | | Human enterovirus 71 3C protease mutant E71A in complex with rupintrivir | | 分子名称: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | 登録日 | 2011-03-07 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.039 Å) | | 主引用文献 | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
3QZQ
 
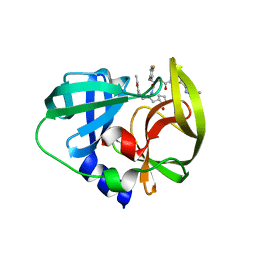 | | Human enterovirus 71 3C protease mutant E71D in complex with rupintrivir | | 分子名称: | 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | 登録日 | 2011-03-07 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7001 Å) | | 主引用文献 | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
2KMK
 
 | | Gfi-1 Zinc Fingers 3-5 complexed with DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*AP*AP*TP*CP*AP*CP*TP*GP*CP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*AP*GP*TP*GP*AP*TP*TP*TP*AP*TP*G)-3'), ZINC ION, ... | | 著者 | Lee, S, Wu, Z. | | 登録日 | 2009-07-30 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Gfi-1 zinc domain bound to consensus DNA.
J.Mol.Biol., 397, 2010
|
|
