1DAQ
 
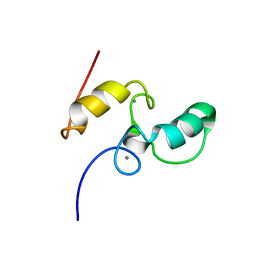 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
4XU6
 
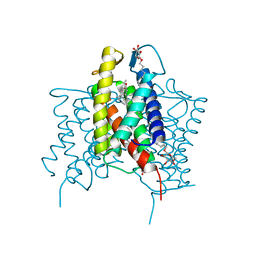 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
5JNX
 
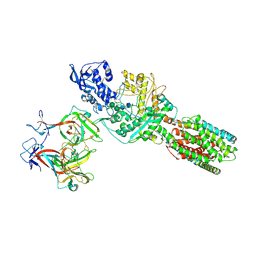 | | The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Wan, T, Shi, Y, Gao, F, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.56 Å) | | Cite: | Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Cell, 165, 2016
|
|
8EZ0
 
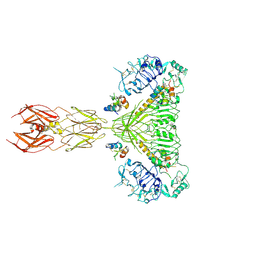 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Symmetric conformation | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYX
 
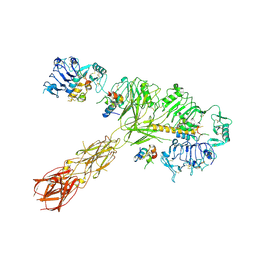 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Asymmetric conformation 1 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYR
 
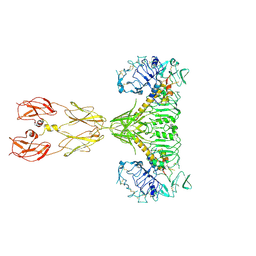 | | Cryo-EM structure of two IGF1 bound full-length mouse IGF1R mutant (four glycine residues inserted in the alpha-CT; IGF1R-P674G4): symmetric conformation | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYY
 
 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S, denoted as IR-3CS) Asymmetric conformation 2 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
5O35
 
 | | Structure of complement proteins complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Complement factor H,Complement factor H | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5IRI
 
 | |
4QVD
 
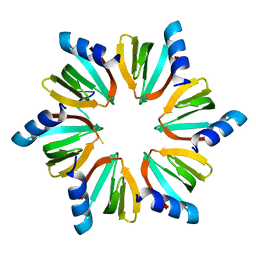 | | E.coli Hfq in complex with RNA Ads | | Descriptor: | RNA (5'-R(*AP*AP*CP*UP*AP*AP*A)-3'), RNA-binding protein Hfq | | Authors: | Wang, L.J, Wang, W.W, Li, F.D, Wu, J.H, Gong, Q.G, Shi, Y.Y. | | Deposit date: | 2014-07-14 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq
Nucleic Acids Res., 43, 2015
|
|
2DK9
 
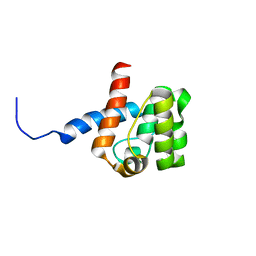 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
6N3I
 
 | |
3OF7
 
 | | The Crystal Structure of Prp20p from Saccharomyces cerevisiae and Its Binding Properties to Gsp1p and Histones | | Descriptor: | Regulator of chromosome condensation | | Authors: | Wu, F, Liu, Y, Zhu, Z, Huang, H, Ding, B, Wu, J, Shi, Y. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9A crystal structure of Prp20p from Saccharomyces cerevisiae and its binding properties to Gsp1p and histones.
J.Struct.Biol., 174, 2011
|
|
7BU5
 
 | | Crystal Structure of Human SLX4 and MUS81 | | Descriptor: | GLYCINE, MUS81 endonuclease homolog (Yeast), isoform CRA_b, ... | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SLX4 control MUS81 endonuclease function in telomere through an intermolecular SAP domain
To be published
|
|
8QPC
 
 | |
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FEI
 
 | | Solution structure of the second SH3 domain of Human CMS protein | | Descriptor: | CD2-associated protein | | Authors: | Yao, B, Dai, H, Jiao, Y, Wu, J, Shi, Y. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH3 domain of human CMS and a newly identified binding site at the C-terminus of c-Cbl
Biochim.Biophys.Acta, 1774, 2007
|
|
3NOA
 
 | |
7YG6
 
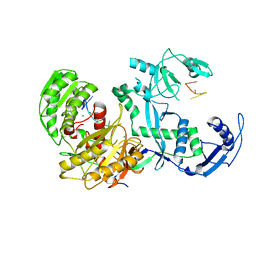 | | Cryo-EM structure of the EfPiwi(N959K) in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YGN
 
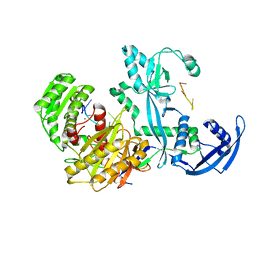 | | Cryo-EM structure of the Mili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFQ
 
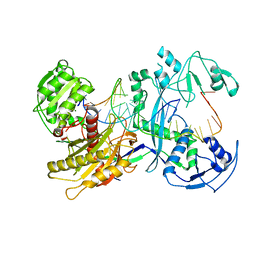 | | Cryo-EM structure of the EfPiwi (N959K)-piRNA-target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi, RNA (5'-R(*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFX
 
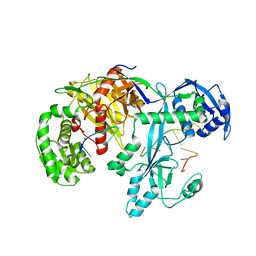 | | Cryo-EM structure of Hili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
5JR7
 
 | | Crystal structure of an ACRDYS heterodimer [RIa(92-365):C] of PKA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Taylor, S.S. | | Deposit date: | 2016-05-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structure of a PKA RI alpha Recurrent Acrodysostosis Mutant Explains Defective cAMP-Dependent Activation.
J. Mol. Biol., 428, 2016
|
|
5H65
 
 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
2DC2
 
 | | Solution Structure of PDZ Domain | | Descriptor: | golgi associated PDZ and coiled-coil motif containing isoform b | | Authors: | Li, X, Wu, J, Shi, Y. | | Deposit date: | 2005-12-20 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GOPC PDZ domain and its interaction with the C-terminal motif of neuroligin
Protein Sci., 15, 2006
|
|
