1G73
 
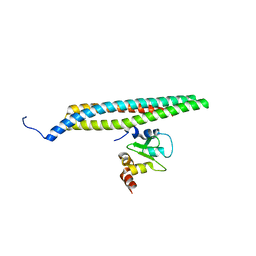 | | CRYSTAL STRUCTURE OF SMAC BOUND TO XIAP-BIR3 DOMAIN | | Descriptor: | INHIBITORS OF APOPTOSIS-LIKE PROTEIN ILP, SECOND MITOCHONDRIA-DERIVED ACTIVATOR OF CASPASES, ZINC ION | | Authors: | Wu, G, Chai, J, Suber, T.L, Wu, J.-W, Shi, Y. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IAP recognition by Smac/DIABLO.
Nature, 408, 2000
|
|
1WM7
 
 | | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch, 9 structures | | Descriptor: | Neurotoxin BmP01 | | Authors: | Wu, G, Li, Y, Wei, D, He, F, Jiang, S, Hu, G, Wu, H, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 276, 2000
|
|
5XQZ
 
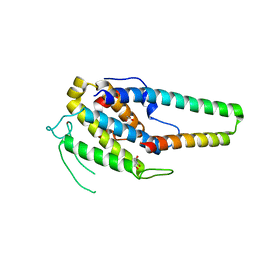 | | Structure of the MOB1-NDR2 complex | | Descriptor: | GLYCEROL, MOB kinase activator 1A, Serine/threonine-protein kinase 38-like, ... | | Authors: | Wu, G, Lin, K. | | Deposit date: | 2017-06-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stable MOB1 interaction with Hippo/MST is not essential for development and tissue growth control.
Nat Commun, 8, 2017
|
|
6Q1U
 
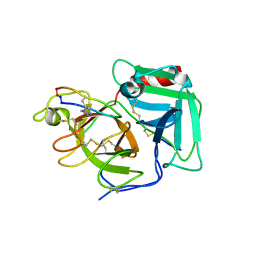 | | Structure of plasmin and peptide complex | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP, Plasminogen | | Authors: | Wu, G, Law, R.H.P. | | Deposit date: | 2019-08-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3467 Å) | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1JIR
 
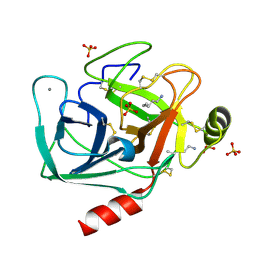 | | Crystal Structure of Trypsin Complex with Amylamine in Cyclohexane | | Descriptor: | AMYLAMINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Wu, G, Zhu, G, Huang, Q, Qian, M, Tang, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of beta-Trypsin Complex with Amylamine in Cyclohexane
To be Published
|
|
1OX1
 
 | | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor | | Descriptor: | 11-mer peptide, CALCIUM ION, Trypsinogen, ... | | Authors: | Wu, G, Huang, Y, Zhu, G, Huang, Q, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-03-31 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor
To be published
|
|
1P22
 
 | | Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity on the SCFbeta-TrCP1 ubiquitin ligase | | Descriptor: | Beta-catenin, F-box/WD-repeat protein 1A, Skp1 | | Authors: | Wu, G, Xu, G, Schulman, B.A, Jeffrey, P.D, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a beta-TrCP1-Skp1-beta-Catenin complex: destruction motif binding and lysine specificity of the SCFbeta-TrCP1 ubiquitin ligase
Mol.Cell, 11, 2003
|
|
2D3H
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 | | Descriptor: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-HYP-HYP-GLY-(PRO-PRO-GLY)4 | | Authors: | Wu, G, Noguchi, K, Okuyama, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2005-09-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
2D3F
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Pro-Hyp-Gly-(Pro-Pro-Gly)4 | | Descriptor: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-PRO-HYP-GLY-(PRO-PRO-GLY)4 | | Authors: | Wu, G, Noguchi, K, Okuyama, K, Ebisuzaki, S, Nishino, N. | | Deposit date: | 2005-09-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
5YK3
 
 | | human Ragulator complex | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Wu, G, Mu, Z. | | Deposit date: | 2017-10-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discov, 3, 2017
|
|
5YK5
 
 | | structure of the human Lamtor4-Lamtor5 complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Wu, G, Mu, Z. | | Deposit date: | 2017-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discov, 3, 2017
|
|
4FQ7
 
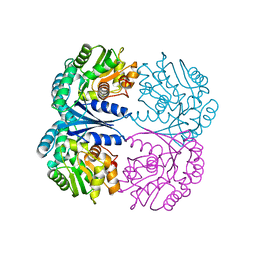 | | Crystal structure of the maleate isomerase Iso from Pseudomonas putida S16 | | Descriptor: | Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
4FQ5
 
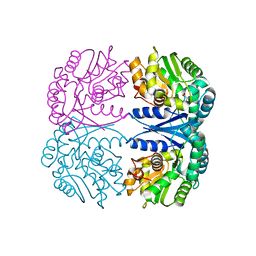 | | Crystal structure of the maleate isomerase Iso(C200A) from Pseudomonas putida S16 with maleate | | Descriptor: | MALEIC ACID, Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
1M2S
 
 | | Solution Structure of A New Potassium Channels Blocker from the Venom of Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Toxin BmTX3 | | Authors: | Wang, Y, Li, M, Zhang, N, Wu, G, Hu, G, Wu, H. | | Deposit date: | 2002-06-25 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The solution structure of BmTx3B, a member of the scorpion toxin subfamily alpha-KTx 16
Proteins, 58, 2005
|
|
1WM8
 
 | | Solution Structure of BmP03 from the Venom of Scorpion Buthus martensii Karsch, 10 structures | | Descriptor: | Neurotoxin BmP03 | | Authors: | Wu, H, He, F, Li, Y, Wu, G, Cao, C, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of BmP03 from Venom of Scorpion Buthus martensii Karsch
Acta Chim.Sinica, 58, 2000
|
|
1WWN
 
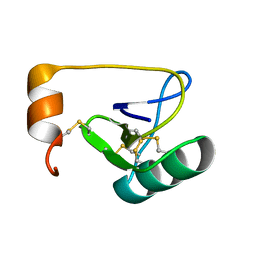 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
7CC9
 
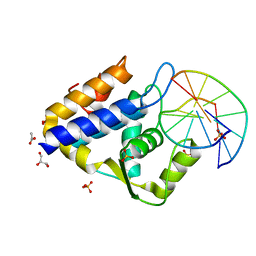 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*GP*CP*GP*GS*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*CP*GP*CP*C)-3'), ... | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCJ
 
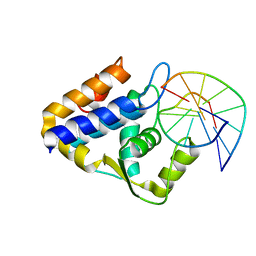 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*AP*TP*C)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
7CMS
 
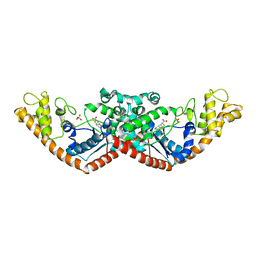 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, PHOSPHATE ION, Tryptophan--tRNA ligase | | Authors: | Lyu, G, Fan, S, Jin, Y, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the specific interaction between Geobacillus stearothermophilus tryptophanyl-tRNA synthetase and antimicrobial Chuangxinmycin.
J.Biol.Chem., 298, 2022
|
|
4OHR
 
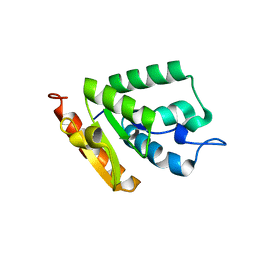 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
2ASS
 
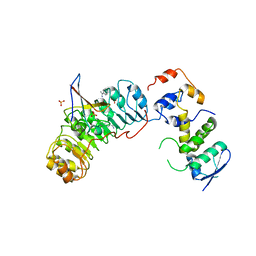 | | Crystal structure of the Skp1-Skp2-Cks1 complex | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinases regulatory subunit 1, PHOSPHATE ION, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
7CKI
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lyu, G, Fan, S, Jin, Y, Liu, J, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP
To Be Published
|
|
