5VMY
 
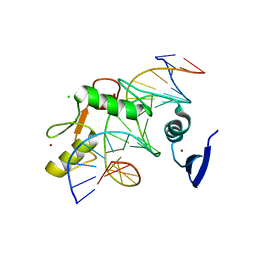 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a hemi CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*CP*GP*CP*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
2AVX
 
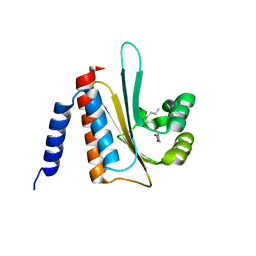 | | solution structure of E coli SdiA1-171 | | Descriptor: | N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE, Regulatory protein sdiA | | Authors: | Yao, Y, Martinez-Yamout, M.A, Dickerson, T.J, Brogan, A.P, Wright, P.E, Dyson, H.J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones.
J.Mol.Biol., 355, 2006
|
|
1R8U
 
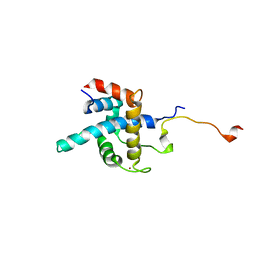 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
2JPA
 
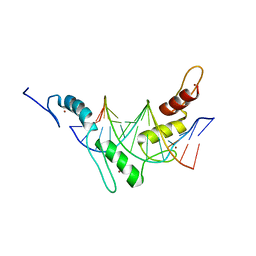 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
1TOT
 
 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
2JP9
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
1SB0
 
 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
1G7O
 
 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
6DMX
 
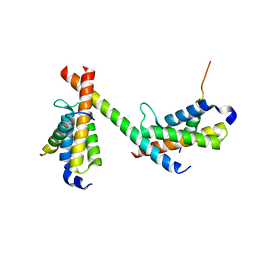 | | HBZ56 in complex with KIX and c-Myb | | Descriptor: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W3Q
 
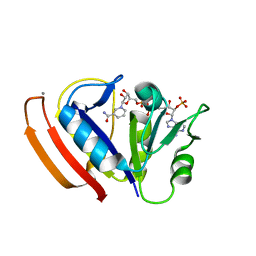 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
2LEF
 
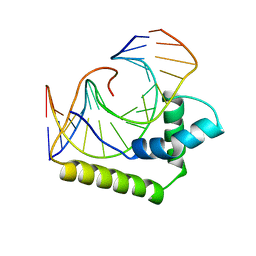 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
5U6H
 
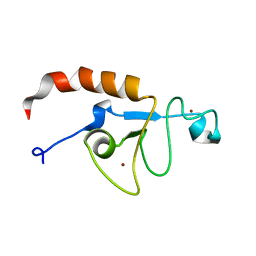 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
5U6L
 
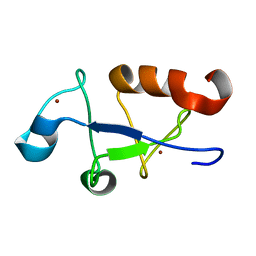 | | Solution structure of the zinc fingers 3 and 4 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
2L14
 
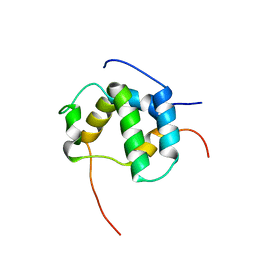 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2LT7
 
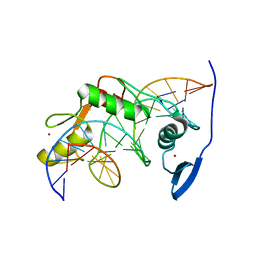 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6DNQ
 
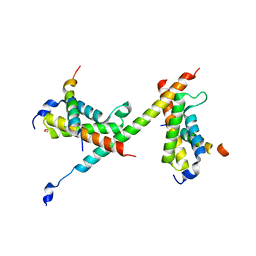 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CC9
 
 | |
5CCC
 
 | |
2AGH
 
 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
2HDP
 
 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
2JOX
 
 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
5U7G
 
 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2KA4
 
 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2KA6
 
 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
2KJE
 
 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | Descriptor: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | Authors: | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
