3GEH
 
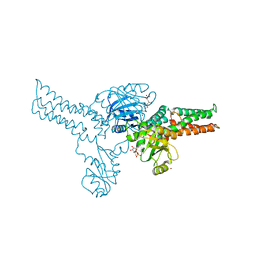 | | Crystal structure of MnmE from Nostoc in complex with GDP, FOLINIC ACID and ZN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ZINC ION, ... | | Authors: | Meyer, S, Wittinghofer, A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Kissing G domains of MnmE monitored by X-ray crystallography and pulse electron paramagnetic resonance spectroscopy
Plos Biol., 7, 2009
|
|
3LVQ
 
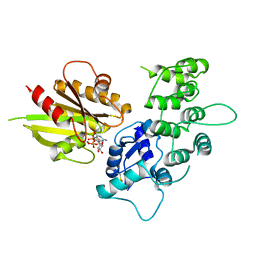 | | The crystal structure of ASAP3 in complex with Arf6 in transition state | | Descriptor: | ALUMINUM FLUORIDE, Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 3, ... | | Authors: | Ismail, S.A, Vetter, I.R, Sot, B, Wittinghofer, A. | | Deposit date: | 2010-02-22 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism
Cell(Cambridge,Mass.), 141, 2010
|
|
3LVR
 
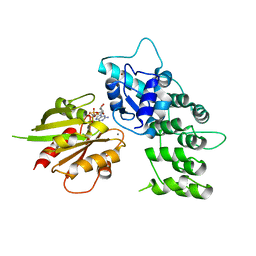 | | The crystal structure of ASAP3 in complex with Arf6 in transition state soaked with Calcium | | Descriptor: | ALUMINUM FLUORIDE, Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 3, ... | | Authors: | Ismail, S.A, Vetter, I.R, Sot, B, Wittinghofer, A. | | Deposit date: | 2010-02-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism
Cell(Cambridge,Mass.), 141, 2010
|
|
5E8F
 
 | | Structure of Fully modified geranylgeranylated PDE6C Peptide in complex with PDE6D | | Descriptor: | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha', GERAN-8-YL GERAN, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Fansa, E.K, O'Reilly, N.J, Ismail, S.A, Wittinghofer, A. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The N- and C-terminal ends of RPGR can bind to PDE6 delta.
Embo Rep., 16, 2015
|
|
5NAL
 
 | | The crystal structure of inhibitor-15 covalently bound to PDE6D | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}1-(cyclohexylmethyl)-~{N}4-cyclopentyl-~{N}1-[(~{Z})-4-[(~{E})-methyliminomethyl]-5-oxidanyl-hex-4-enyl]benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent Protein Labeling at Glutamic Acids.
Cell Chem Biol, 24, 2017
|
|
2NTY
 
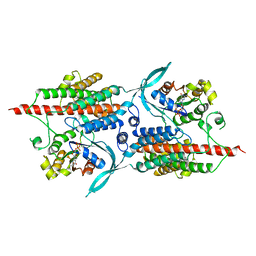 | | Rop4-GDP-PRONE8 | | Descriptor: | Emb|CAB41934.1, GUANOSINE-5'-DIPHOSPHATE, Rac-like GTP-binding protein ARAC5 | | Authors: | Thomas, C, Fricke, I, Scrima, A, Berken, A, Wittinghofer, A. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Evidence for a Common Intermediate in Small G Protein-GEF Reactions
Mol.Cell, 25, 2007
|
|
3I3S
 
 | | Crystal Structure of H-Ras with Thr50 replaced by Isoleucine | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gremer, L, Dvorsky, R, Merbitz-Zahradnik, T, Wittinghofer, A, Ahmadian, M.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A restricted spectrum of NRAS mutations causes Noonan syndrome.
Nat.Genet., 42, 2010
|
|
2NTX
 
 | | Prone8 | | Descriptor: | Emb|CAB41934.1 | | Authors: | Thomas, C, Fricke, I, Scrima, A, Berken, A, Wittinghofer, A. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Evidence for a Common Intermediate in Small G Protein-GEF Reactions
Mol.Cell, 25, 2007
|
|
5ML4
 
 | | The crystal structure of PDE6D in complex to inhibitor-7 | | Descriptor: | 4-[[[4-[(4-chlorophenyl)methyl-cyclopentyl-sulfamoyl]phenyl]sulfonyl-(piperidin-4-ylmethyl)amino]methyl]-2-(methylamino)benzoic acid, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Fansa, E.K, Martin-Gago, P, waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML2
 
 | | The crystal structure of PDE6D in complex with inhibitor-3 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}1-[(4-chlorophenyl)methyl]-~{N}1-cyclopentyl-~{N}4-(phenylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML8
 
 | | The crystal structure of PDE6D in complex to inhibitor-4 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}4-cyclopentyl-~{N}1-(phenylmethyl)-~{N}1-(piperidin-4-ylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4ZI3
 
 | | BART-like domain of BARTL1/CCDC104 aa1-133 in complex with Arl3FL bound to GppNHp in P1 21 1 | | Descriptor: | ADP-ribosylation factor-like protein 3, Cilia- and flagella-associated protein 36, MAGNESIUM ION, ... | | Authors: | Lokaj, M, Koerner, C, Koesling, S, Wittinghofer, A. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Interaction of CCDC104/BARTL1 with Arl3 and Implications for Ciliary Function.
Structure, 23, 2015
|
|
4ZI2
 
 | | BART-like domain of BARTL1/CCDC104 in complex with Arl3FL bound to GppNHp in P21 21 21 | | Descriptor: | ADP-ribosylation factor-like protein 3, Cilia- and flagella-associated protein 36, MAGNESIUM ION, ... | | Authors: | Lokaj, M, Koerner, C, Koesling, S, Wittinghofer, A. | | Deposit date: | 2015-04-27 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Interaction of CCDC104/BARTL1 with Arl3 and Implications for Ciliary Function.
Structure, 23, 2015
|
|
5ML6
 
 | | The crystal structure of PDE6D in complex to inhibitor-8 | | Descriptor: | 2-azanyl-4-[[[4-[(4-chlorophenyl)methyl-cyclopentyl-sulfamoyl]phenyl]sulfonyl-(piperidin-4-ylmethyl)amino]methyl]benzoic acid, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Fansa, E.K, Martin-gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML3
 
 | | The crystal structure of PDE6D in complex to Deltasonamide1 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}1-[(4-chlorophenyl)methyl]-~{N}1-cyclopentyl-~{N}4-[[2-(methylamino)pyrimidin-4-yl]methyl]-~{N}4-(piperidin-4-ylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5E80
 
 | | The crystal structure of PDEd in complex with inhibitor-2a | | Descriptor: | N-(3-chloro-2-methylphenyl)-4-(3,4-dimethyl-7-oxo-2-phenyl-2,7-dihydro-6H-pyrazolo[3,4-d]pyridazin-6-yl)butanamide, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Ismail, S, Fansa, E.K, Murarka, S, Wittinghofer, A. | | Deposit date: | 2015-10-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of pyrazolopyridazinones as PDE delta inhibitors.
Nat Commun, 7, 2016
|
|
5DI3
 
 | | Crystal structure of Arl13B in complex with Arl3 of Chlamydomonas reinhardtii | | Descriptor: | ADP-ribosylation factor-like protein 13B, ADP-ribosylation factor-like protein 3, MAGNESIUM ION, ... | | Authors: | Gotthardt, K, Lokaj, M, Falk, N, Koerner, C, Giessl, A, Wittinghofer, A. | | Deposit date: | 2015-08-31 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins.
Elife, 4, 2015
|
|
4YZM
 
 | | Humanized Roco4 bound to LRRK2-In1 | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, MAGNESIUM ION, Probable serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Messias, A.C, Ito, G, Sattler, M, Alessi, D.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of LRRK2 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YZN
 
 | | Humanized Roco4 bound to Compound 19 | | Descriptor: | (4-{[4-(cyclopropylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-2-fluoro-5-methoxyphenyl)(morpholin-4-yl)methanone, Probable serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Messias, A.C, Ito, G, Sattler, M, Alessi, D.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of LRRK2 Inhibitors.
J.Med.Chem., 58, 2015
|
|
2D4H
 
 | | Crystal-structure of the N-terminal large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1 | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
1A4R
 
 | | G12V MUTANT OF HUMAN PLACENTAL CDC42 GTPASE IN THE GDP FORM | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, G25K GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rudolph, M.G, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 1998-02-02 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleotide binding to the G12V-mutant of Cdc42 investigated by X-ray diffraction and fluorescence spectroscopy: two different nucleotide states in one crystal.
Protein Sci., 8, 1999
|
|
1K5G
 
 | | Crystal structure of Ran-GDP-AlFx-RanBP1-RanGAP complex | | Descriptor: | ALUMINUM FLUORIDE, GTP-binding nuclear protein RAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
2B8W
 
 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP/AlF4 | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2B92
 
 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GDP/AlF3 | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Interferon-induced guanylate-binding protein 1, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
1K5D
 
 | | Crystal structure of Ran-GPPNHP-RanBP1-RanGAP complex | | Descriptor: | GTP-binding nuclear protein RAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
