4N5K
 
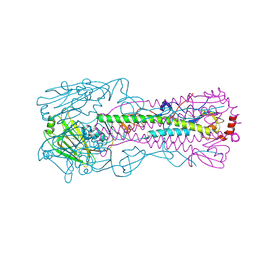 | |
4NZR
 
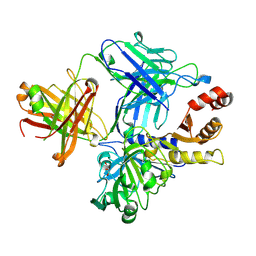 | |
4NM8
 
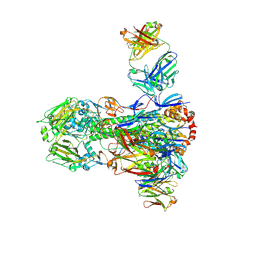 | |
4NZT
 
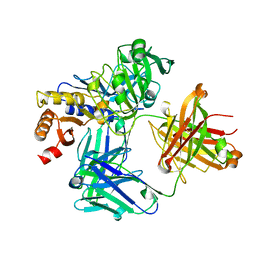 | |
4NM4
 
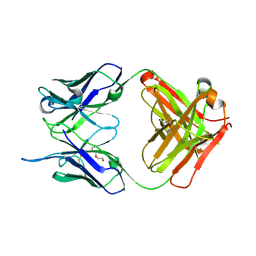 | |
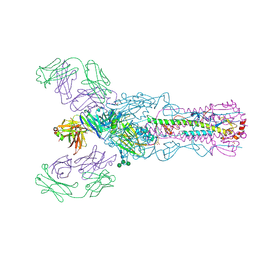
4O58
 
 | |
4NUJ
 
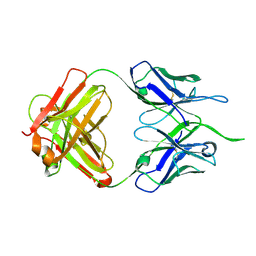 | |
4NZU
 
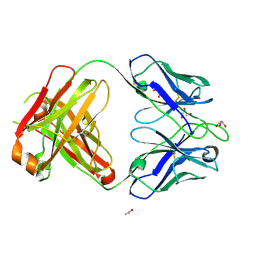 | |
4NWT
 
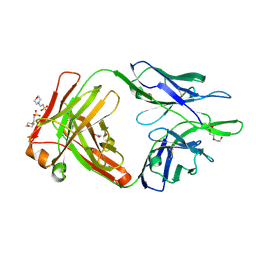 | | Crystal structure of the anti-human NGF Fab APE1531 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, APE1531 Ab Fab heavy chain, ... | | Authors: | Verdino, P, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2013-12-06 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nucleotide insertions and deletions complement point mutations to massively expand the diversity created by somatic hypermutation of antibodies.
J.Biol.Chem., 289, 2014
|
|
1GGB
 
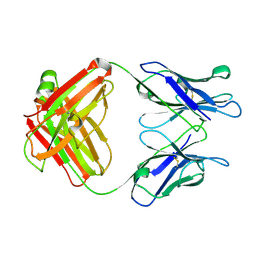 | |
1GGC
 
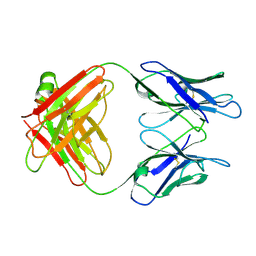 | |
1GGI
 
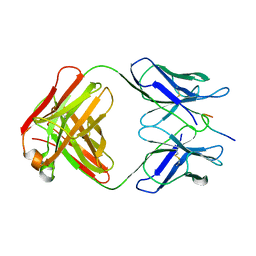 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6KVF
 
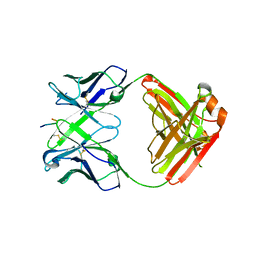 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
6KYZ
 
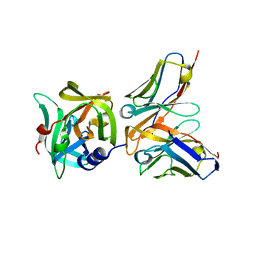 | | HRV14 3C in complex with single chain antibody YDF | | Descriptor: | Genome polyprotein, YDF H chain, YDF L chain | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M87
 
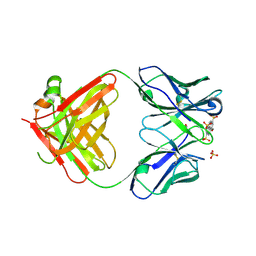 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6ME1
 
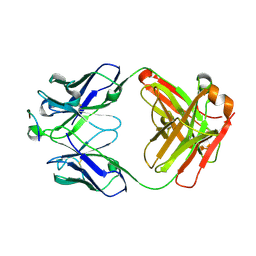 | |
6MG4
 
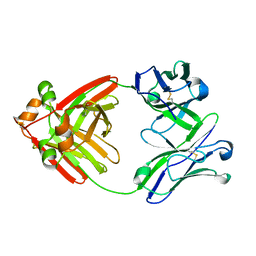 | | Structure of full-length human lambda-6A light chain JTO | | Descriptor: | JTO light chain | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MCO
 
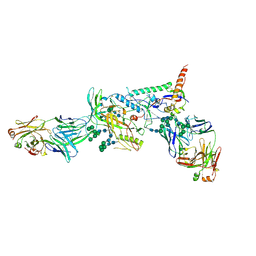 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P23 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-08-31 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6MG5
 
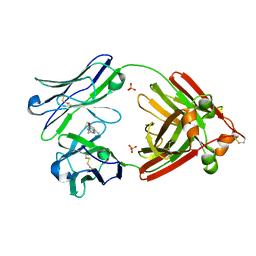 | | Structure of full-length human lambda-6A light chain JTO in complex with coumarin 1 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, Light chain JTO, PHOSPHATE ION | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MDT
 
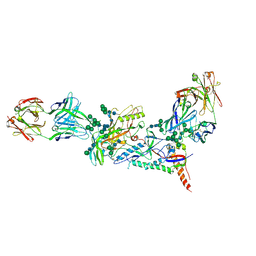 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P63 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.816 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6MSY
 
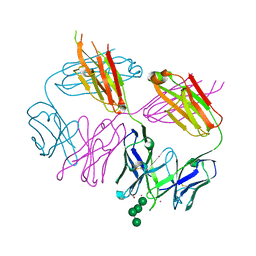 | | Anti-HIV-1 Fab Fab 2G12 + Man4 re-refinement | | Descriptor: | ACETATE ION, Fab 2G12, light chain, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6N2X
 
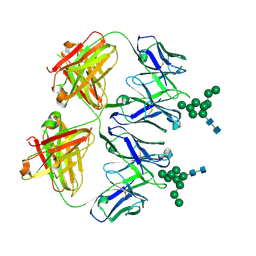 | | Anti-HIV-1 Fab 2G12 + Man9 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
6MNF
 
 | | Anti-HIV-1 Fab 2G12 + Man8 re-refinement | | Descriptor: | Fab 2G12, light chain, Fab 2g12, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
1IAO
 
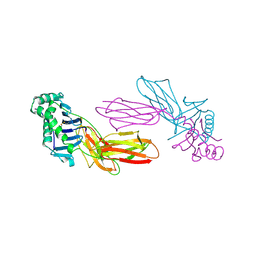 | | CLASS II MHC I-AD IN COMPLEX WITH OVALBUMIN PEPTIDE 323-339 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-AD | | Authors: | Scott, C.A, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 1998-03-13 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues.
Immunity, 8, 1998
|
|
6N35
 
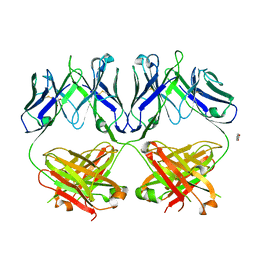 | | Anti-HIV-1 Fab 2G12 + Man1-2 re-refinement | | Descriptor: | BENZOIC ACID, Fab 2G12 heavy chain, Fab 2G12 light chain, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
