5ABB
 
 | | Visualization of a polytopic membrane protein during SecY-mediated membrane insertion | | Descriptor: | GREEN-LIGHT ABSORBING PROTEORHODOPSIN, PROTEIN TRANSLOCASE SUBUNIT SECE, PROTEIN TRANSLOCASE SUBUNIT SECY | | Authors: | Bischoff, L, Wickles, S, Berninghausen, O, vanderSluis, E, Beckmann, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Nat.Commun., 5, 2014
|
|
7NFC
 
 | | Cryo-EM structure of NHEJ super-complex (dimer) | | Descriptor: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
 | | Cryo-EM structure of NHEJ super-complex (monomer) | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
2YJV
 
 | |
7AUA
 
 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) in complex with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
7AU2
 
 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
6GWK
 
 | | The crystal structure of Hfq from Caulobacter crescentus | | Descriptor: | RNA-binding protein Hfq | | Authors: | Santiago-Frangos, A, Frohlich, K.S, Jeliazkov, J.R, Gray, J.R, Luisi, B.F, Woodson, S.A, Hardwick, S.W. | | Deposit date: | 2018-06-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Caulobacter crescentusHfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4DKF
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb2 | | Descriptor: | FAb2 Heavy Chain, FAb2 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
4DKE
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb1.1 | | Descriptor: | FAb1.1 Heavy Chain, FAb1.1 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
7PCH
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:2 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCF
 
 | | Human methemoglobin bound to Staphylococcus aureus hemophore IsdB | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.82 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCQ
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:1 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QNA
 
 | | Cryo-EM structure of human full-length alpha4beta3gamma2 GABA(A)R in complex with GABA and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNB
 
 | | Cryo-EM structure of human full-length beta3gamma2 GABA(A)R in complex with GABA and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7B1D
 
 | | Cryo-EM of Aedes Aegypti Toll5A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7B1B
 
 | | Cryo-EM of Aedes Aegypti Toll5A dimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
9H9E
 
 | | Cryo-EM structure of the human GABAA receptor alpha1 subunit in complex with the assembly factor NACHO/TMEM35A | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Hooda, Y, Sente, A, Judy, R.M, Smalinskaite, L, Peak-Chew, S, Naydenova, K, Malinauskas, T, Hardwick, S.W, Chirgadze, D.Y, Aricescu, A.R, Hegde, R.S. | | Deposit date: | 2024-10-30 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of NACHO-mediated assembly of pentameric ligand-gated ion channels.
Biorxiv, 2024
|
|
5A7U
 
 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
6ZFP
 
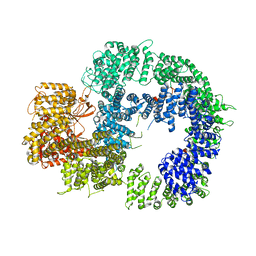 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
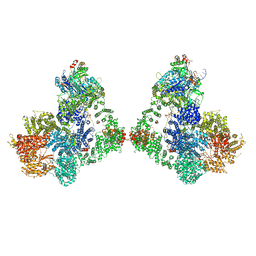 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
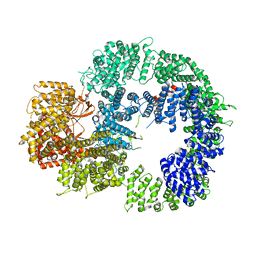 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
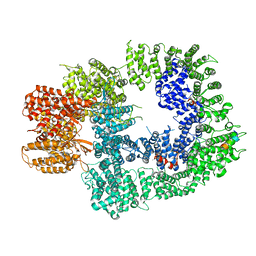 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7QNC
 
 | | Cryo-EM structure of human full-length extrasynaptic alpha4beta3delta GABA(A)R in complex with THIP (gaboxadol), histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
