2AVQ
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
1AGX
 
 | | REFINED CRYSTAL STRUCTURE OF ACINETOBACTER GLUTAMINASIFICANS GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Housset, D, Weber, I.T, Ammon, H.L, Murphy, K.C, Swain, A.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of Acinetobacter glutaminasificans glutaminase-asparaginase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
8F0F
 
 | | HIV-1 wild type protease with GRL-110-19A, a chloroacetamide derivative based on Darunavir as P2' group | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[4-(2-chloroacetamido)benzene-1-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.
Bioorg.Med.Chem.Lett., 83, 2023
|
|
1A8M
 
 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | Descriptor: | TUMOR NECROSIS FACTOR ALPHA | | Authors: | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | Deposit date: | 1998-03-27 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
8FUI
 
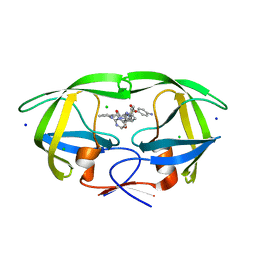 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
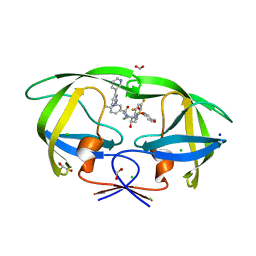 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
5JG1
 
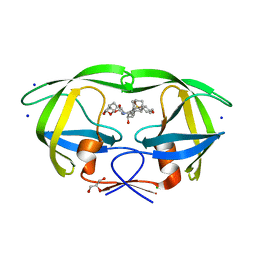 | | HIV-1 wild Type protease with GRL-031-14A (a Adamantane P1-Ligand with tetrahydropyrano-tetrahydrofuran in P2 and isobutylamine in P1') | | Descriptor: | (3R,3aS,7aR)-hexahydro-4H-furo[2,3-b]pyran-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
3D1Y
 
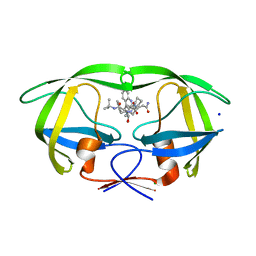 | | Crystal structure of HIV-1 mutant I54V and inhibitor SAQUINA | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3CYX
 
 | | Crystal structure of HIV-1 mutant I50V and inhibitor saquinavira | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D1Z
 
 | | Crystal structure of HIV-1 mutant I54M and inhibitor DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Liu, F, Kovalesky, A.Y, Tie, Y, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2008-05-07 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D20
 
 | | Crystal structure of HIV-1 mutant I54V and inhibitor DARUNAVIA | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Liu, F, Kovalesky, A.Y, Tie, Y, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2008-05-07 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D1X
 
 | | Crystal structure of HIV-1 mutant I54M and inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3CYW
 
 | | Effect of Flap Mutations on Structure of HIV-1 Protease and Inhibition by Saquinavir and Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
7TO5
 
 | | HIV-1 wild type protease with GRL-05816A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 1] | | Descriptor: | (1R,3aS,4S,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
7TO6
 
 | | HIV-1 wild type protease with GRL-01717A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 2] | | Descriptor: | (1S,3aR,4R,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
5UOV
 
 | | HIV-1 wild Type protease with GRL-1118A , an isophthalamide-derived P2-P3 ligand with the sulfonamide isostere as the P2' group | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
5ULT
 
 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
5UPZ
 
 | | HIV-1 wild Type protease with GRL-0518A , an isophthalamide-derived P2-P3 ligand with the para-hydoxymethyl sulfonamide isostere as the P2' group | | Descriptor: | CHLORIDE ION, GLYCEROL, N~3~-{(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}-N~1~-methyl-N~1~-[(4-methyl-1,3-oxazol-2-yl)methyl]benzene-1,3-dicarboxamide, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
4NPU
 
 | |
4NPT
 
 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
4Q4K
 
 | | Crystal structure of nitronate monooxygenase from Pseudomonas aeruginosa PAO1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitronate Monooxygenase | | Authors: | Salvi, F, Agniswamy, J, Gadda, G, Weber, I.T. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Combined Structural and Kinetic Characterization of a Bacterial Nitronate Monooxygenase from Pseudomonas aeruginosa PAO1 Establishes NMO Class I and II.
J.Biol.Chem., 289, 2014
|
|
6U7O
 
 | | HIV-1 wild type protease with GRL-00819A, with phenyl-boronic-acid as P2'-ligand and with a 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
6U7P
 
 | | HIV-1 wild type protease with GRL-03119A, with phenyl-boronic-acid as P2'-ligand and with a hexahydro-4H-furo-pyran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
6VCE
 
 | | HIV-1 wild type protease with GRL-026-18A, a crown-like tetrahydropyranotetrahydrofuran with a bridged methylene group as a P2 ligand | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]-2-[(3S,3aR,5S,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl]acetamide, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Design, Synthesis, and X-ray Studies of Potent HIV-1 Protease Inhibitors with P2-Carboxamide Functionalities.
Acs Med.Chem.Lett., 11, 2020
|
|
