8JDH
 
 | |
3OXZ
 
 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | 分子名称: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | 著者 | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | 登録日 | 2010-09-22 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
4EYR
 
 | | Crystal structure of multidrug-resistant clinical isolate 769 HIV-1 protease in complex with ritonavir | | 分子名称: | HIV-1 PROTEASE, RITONAVIR | | 著者 | Liu, Z, Yedidi, R.S, Wang, Y, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | 登録日 | 2012-05-01 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into the mechanism of drug resistance: X-ray structure analysis of multi-drug resistant HIV-1 protease ritonavir complex.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
3OU4
 
 | | MDR769 HIV-1 protease complexed with TF/PR hepta-peptide | | 分子名称: | HIV-1 protease, TF/PR substrate peptide | | 著者 | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | 登録日 | 2010-09-14 | | 公開日 | 2011-03-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OU3
 
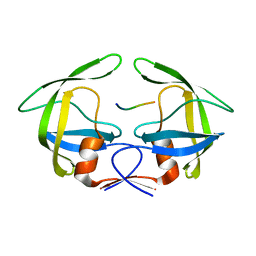 | | MDR769 HIV-1 protease complexed with PR/RT hepta-peptide | | 分子名称: | HIV-1 protease, PR/RT substrate peptide | | 著者 | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | 登録日 | 2010-09-14 | | 公開日 | 2011-03-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUC
 
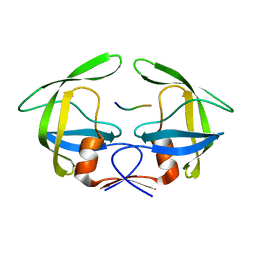 | | MDR769 HIV-1 protease complexed with p2/NC hepta-peptide | | 分子名称: | MDR HIV-1 protease, p2/NC substrate peptide | | 著者 | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | 登録日 | 2010-09-14 | | 公開日 | 2011-03-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
4GYE
 
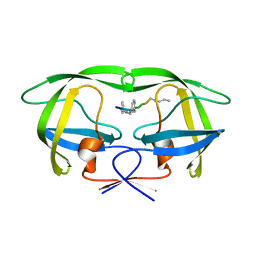 | | MDR 769 HIV-1 Protease in Complex with Reduced P1F | | 分子名称: | P1F peptide, Protease | | 著者 | Dewdney, T.G, Wang, Y, Brunzelle, J, Reiter, S.J, Kovari, I.A, Kovari, L.C. | | 登録日 | 2012-09-05 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
5WQN
 
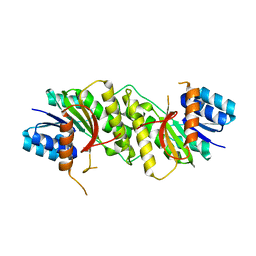 | |
5WQP
 
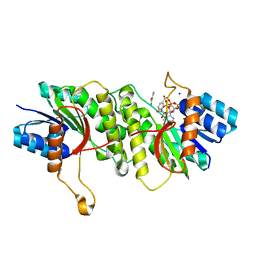 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | 著者 | Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2016-11-27 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | 分子名称: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | 著者 | Yang, J, Wang, Y, Zhang, G. | | 登録日 | 2023-03-01 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
3OE7
 
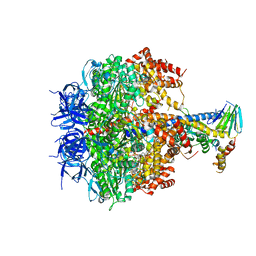 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | 分子名称: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | 著者 | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | 登録日 | 2010-08-12 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3OAI
 
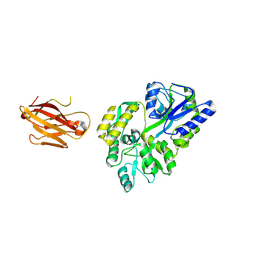 | | Crystal structure of the extra-cellular domain of human myelin protein zero | | 分子名称: | Maltose-binding periplasmic protein, Myelin protein P0, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Sohi, J, Kamholz, J, Kovari, L.C. | | 登録日 | 2010-08-05 | | 公開日 | 2011-12-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the extracellular domain of human myelin protein zero.
Proteins, 80, 2012
|
|
4EKU
 
 | |
3OEE
 
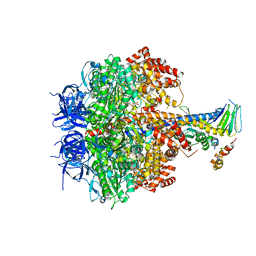 | | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S | | 分子名称: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | 著者 | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | 登録日 | 2010-08-12 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S
To be Published
|
|
3OFN
 
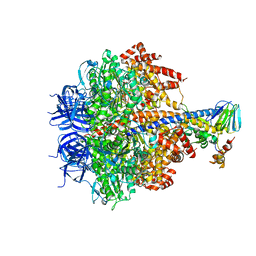 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | 分子名称: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | 著者 | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | 登録日 | 2010-08-15 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
4E6K
 
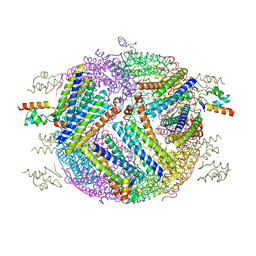 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | 分子名称: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | 著者 | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | 登録日 | 2012-03-15 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
5WQO
 
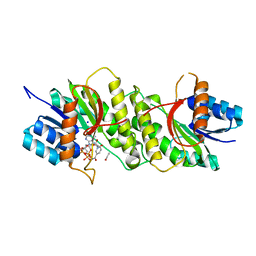 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | 分子名称: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | 著者 | Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2016-11-27 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
2F0X
 
 | | Crystal structure and function of human thioesterase superfamily member 2(THEM2) | | 分子名称: | SULFATE ION, Thioesterase superfamily member 2 | | 著者 | Cheng, Z, Song, F, Shan, X, Wang, Y, Wei, Z, Gong, W. | | 登録日 | 2005-11-14 | | 公開日 | 2006-10-10 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of human thioesterase superfamily member 2
Biochem.Biophys.Res.Commun., 349, 2006
|
|
5WQM
 
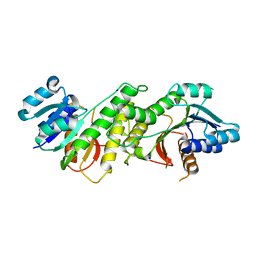 | |
4GZF
 
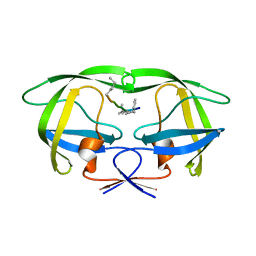 | | Multi-drug resistant HIV-1 protease 769 variant with reduced LrF peptide | | 分子名称: | LrF peptide, Protease | | 著者 | Dewdney, T.G, Wang, Y, Kovari, I.A, Brunzelle, J.S, Reiter, S.J, Kovari, L.C. | | 登録日 | 2012-09-06 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
3B96
 
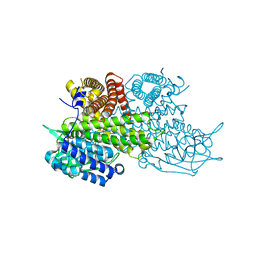 | | Structural Basis for Substrate Fatty-Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, TETRADECANOYL-COA, Very long-chain specific acyl-CoA dehydrogenase | | 著者 | McAndrew, R.P, Wang, Y, Mohsen, A.W, He, M, Vockley, J, Kim, J.J. | | 登録日 | 2007-11-02 | | 公開日 | 2008-02-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3PR2
 
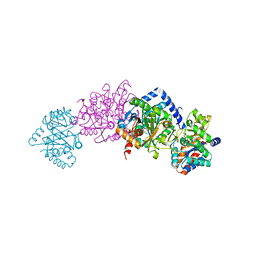 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | 分子名称: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | 著者 | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | 登録日 | 2010-11-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
2JJ3
 
 | | Estrogen receptor beta ligand binding domain in complex with a Benzopyran agonist | | 分子名称: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, ESTROGEN RECEPTOR BETA | | 著者 | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S, Reilly, J.E, Ryter, K.T. | | 登録日 | 2007-07-03 | | 公開日 | 2007-08-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Benzopyrans as Selective Estrogen Receptor Beta Agonists (Serbas). Part 4: Functionalization of the Benzopyran A-Ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | 著者 | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | 登録日 | 2020-06-08 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-09-30 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | 分子名称: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | 著者 | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | 登録日 | 2020-06-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
