2QCI
 
 | | HIV-1 Protease mutant D30N with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD8
 
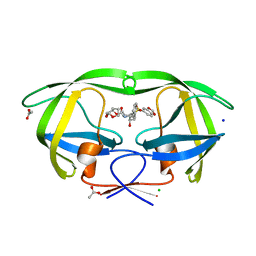 | | HIV-1 Protease Mutant I84V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD7
 
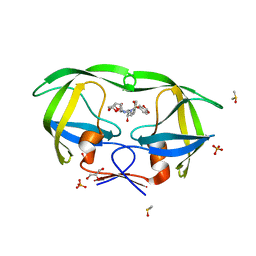 | | HIV-1 Protease Mutant V82A with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD6
 
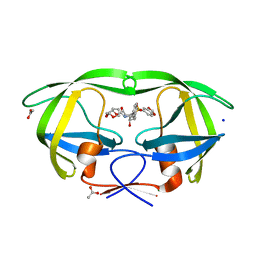 | | HIV-1 Protease Mutant I50V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2Z4O
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
7WD0
 
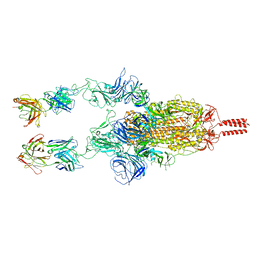 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
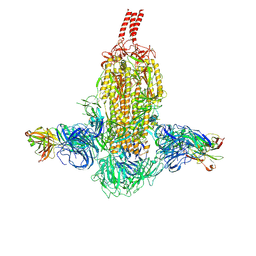 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD7
 
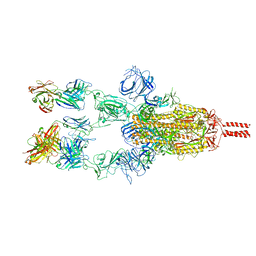 | | SARS-CoV-2 Beta spike in complex with three S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCZ
 
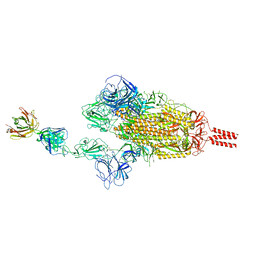 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
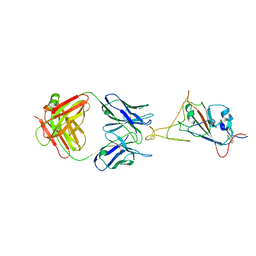 | | RBD-1 of SARS-CoV-2 Beta spike in complex with S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD8
 
 | | SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
3DK1
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0105A | | Descriptor: | (4aR,6r,7aS)-hexahydro-4aH-cyclopenta[b][1,4]dioxin-6-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies
Org.Biomol.Chem., 6, 2008
|
|
3DJK
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0255A | | Descriptor: | (5R)-1,3-dioxepan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-23 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies
J.Med.Chem., 51, 2008
|
|
1EBK
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1JNP
 
 | | Crystal Structure of Murine Tcl1 at 2.5 Resolution | | Descriptor: | T-CELL LEUKEMIA/LYMPHOMA PROTEIN 1A | | Authors: | Petock, J.M, Torshin, I.Y, Wang, Y.F, DuBois, G.C, Croce, C.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2001-07-24 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of murine Tcl1 at 2.5 A resolution and implications for the TCL oncogene family.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7DK4
 
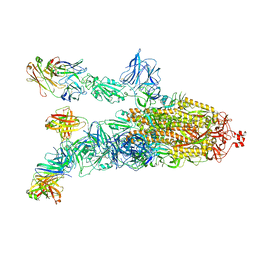 | |
6DKN
 
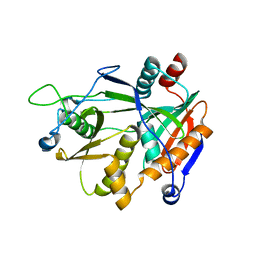 | |
8YO8
 
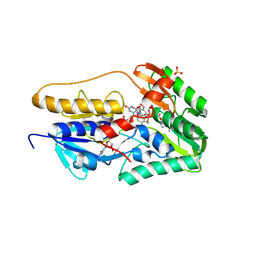 | | Crystal structure of hFSP1 with both 6-OH-FAD and NAD (hFSP1-6-OH-FAD-NAD) | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lan, H.Y, Gao, Y, Hong, T, Zhao, Z.Z, Chang, Z.H, Wang, Y.F, Wang, F. | | Deposit date: | 2024-03-12 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into 6-OH-FAD-dependent activation of hFSP1 for ferroptosis suppression.
Cell Discov, 10, 2024
|
|
8YOQ
 
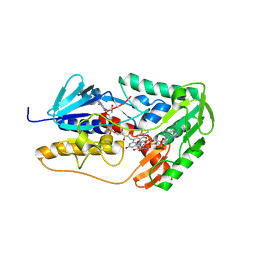 | | Crystal structure of hFSP1 with both 6-OH-FAD and NADP (hFSP1-6-OH-FAD-NADP) | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lan, H.Y, Gao, Y, Hong, T, Zhao, Z.Z, Chang, Z.H, Wang, Y.F, Wang, F. | | Deposit date: | 2024-03-13 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into 6-OH-FAD-dependent activation of hFSP1 for ferroptosis suppression.
Cell Discov, 10, 2024
|
|
8YOX
 
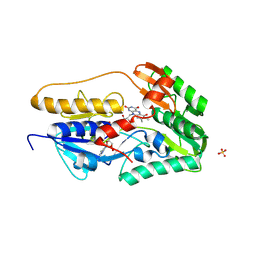 | | Crystal structure of hFSP1 with 6-OH-FAD (hFSP1-6-OH-FAD) | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, SULFATE ION | | Authors: | Lan, H.Y, Gao, Y, Hong, T, Zhao, Z.Z, Chang, Z.H, Wang, Y.F, Wang, F. | | Deposit date: | 2024-03-14 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insight into 6-OH-FAD-dependent activation of hFSP1 for ferroptosis suppression.
Cell Discov, 10, 2024
|
|
1TQH
 
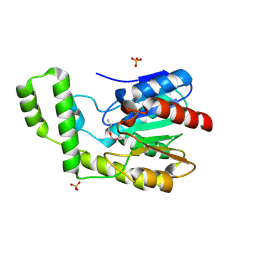 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
7YS6
 
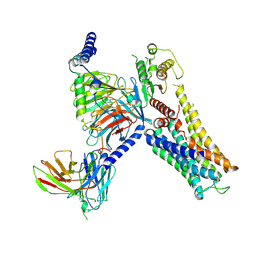 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1DAZ
 
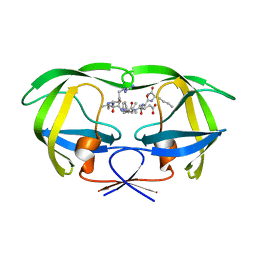 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE (RETROPEPSIN), N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1DW6
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
