5H6Y
 
 | |
4PFC
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-(5-{5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}-2-fluorophenyl)-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
5G4C
 
 | | Human SIRT2 catalyse short chain fatty acyl lysine | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, SIRT2, ... | | Authors: | Wang, Y. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones.
J. Am. Chem. Soc., 138, 2016
|
|
7X7D
 
 | | SARS-CoV-2 Delta RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1 | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
7X7E
 
 | | SARS-CoV-2 RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
8HK5
 
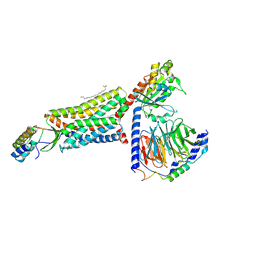 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
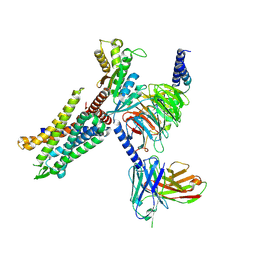 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK2
 
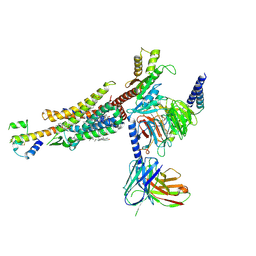 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
7K6B
 
 | |
7KRB
 
 | |
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQR
 
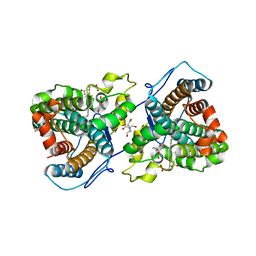 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
8HYA
 
 | |
8XAU
 
 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAV
 
 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
5XIW
 
 | | Crystal structure of T2R-TTL-Colchicine complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-04-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
3SPK
 
 | | Tipranavir in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE | | Authors: | Wang, Y, Liu, Z, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The higher barrier of darunavir and tipranavir resistance for HIV-1 protease.
Biochem.Biophys.Res.Commun., 412, 2011
|
|
5XP3
 
 | | Crystal structure of apo T2R-TTL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
3V74
 
 | | crystal structure of FBF-2 in complex with gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*CP*AP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Wang, Y, Qiu, C, Kershner, A, Holley, C.P, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
7W5E
 
 | | Oxidase ChaP D49L mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Wang, Y, Zheng, W, Meng, Z, Jin, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3ZG6
 
 | | The novel de-long chain fatty acid function of human sirt6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-6, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2012-12-15 | | Release date: | 2013-04-03 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sirt6 Regulates Tnf-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine
Nature, 496, 2013
|
|
2KKR
 
 | | Solution structure of SCA7 zinc finger domain from human ataxin-7 protein | | Descriptor: | Ataxin-7, ZINC ION | | Authors: | Wang, Y, Atkinson, A.R, Bonnet, J, Romier, C, Kieffer, B, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Histone deubiquitination by SAGA is modulated by an atypical zinc finger domain of Ataxin-7
To be Published
|
|
