7WC8
 
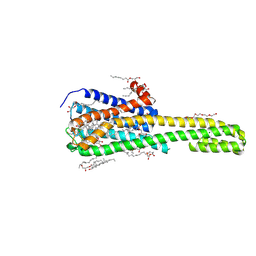 | | Crystal structure of serotonin 2A receptor in complex with lumateperone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(10~{R},15~{S})-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5,7,9(16)-trien-12-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
3CWU
 
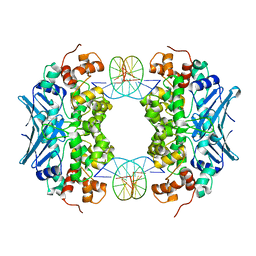 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxy-1,N6-ethenoadenine:Thymine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FE)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DTP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CWS
 
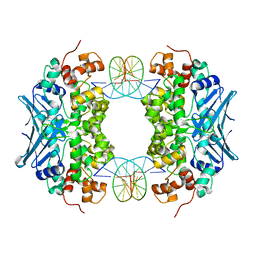 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxyinosine:Thymine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FI)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DTP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CWA
 
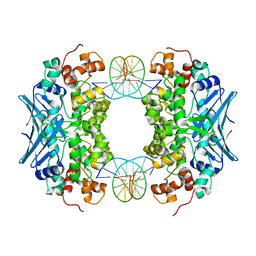 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*(8OG)P*DAP*DCP*DAP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CVT
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*(8OG)P*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3N99
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, uncharacterized protein TM1086 | | Authors: | Chruszcz, M, Domagalski, M.J, Wang, S, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
1OR7
 
 | | Crystal Structure of Escherichia coli sigmaE with the Cytoplasmic Domain of its Anti-sigma RseA | | Descriptor: | RNA polymerase sigma-E factor, Sigma-E factor negative regulatory protein | | Authors: | Campbell, E.A, Tupy, J.L, Gruber, T.M, Wang, S, Sharp, M.M, Gross, C.A, Darst, S.A. | | Deposit date: | 2003-03-12 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA.
Mol.Cell, 11, 2003
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
4GM3
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-101 | | Descriptor: | MM-101, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.393 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
3CVS
 
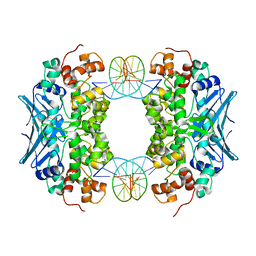 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3ROV
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
3D4V
 
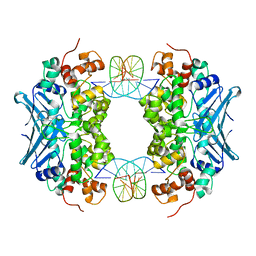 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
4GM9
 
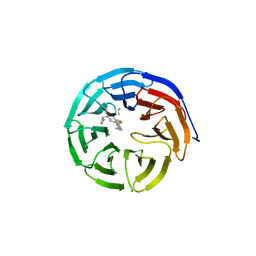 | | Crystal structure of human WD repeat domain 5 with compound MM-401 | | Descriptor: | MM-401, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Liu, L, Dou, Y, Lei, M, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human WD repeat domain 5 with compound MM-401
To be Published
|
|
9FGO
 
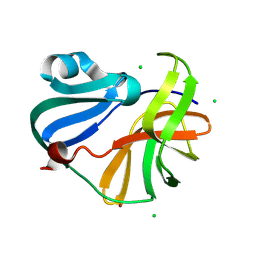 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
9BW1
 
 | | TnsABCD-DNA transpososome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Integrase, LE_polyA, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of TnsABCD transpososome reveals mechanisms of targeted DNA transposition.
Cell, 2024
|
|
4Y49
 
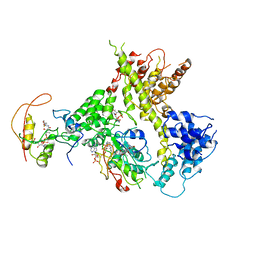 | |
7Y1B
 
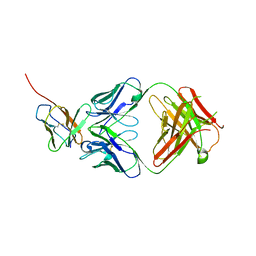 | | 3.2 angstrom cryo-EM structure of extracellular region of mouse Basigin-2 in complex with the Fab fragment of antibody 6E7F1 | | Descriptor: | Heavy chain of 6E7F1, Isoform 2 of Basigin, Light chain of 6E7F1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin
To Be Published
|
|
2LWG
 
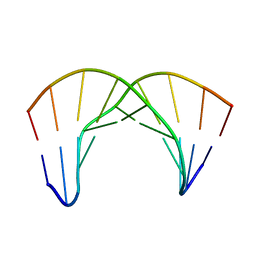 | |
2LWH
 
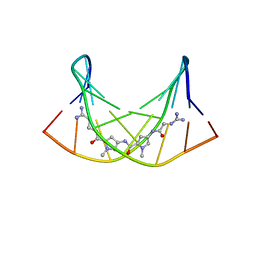 | | NMR Structure of the Self-Complementary 10 mer DNA Duplex 5'-GGATATATCC-3' in Complex with Netropsin | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*AP*TP*AP*TP*CP*C)-3'), NETROPSIN | | Authors: | Rettig, M, Germann, M.W, Wilson, W, Wang, S. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for sequence-dependent induced DNA bending.
Chembiochem, 14, 2013
|
|
4ZNS
 
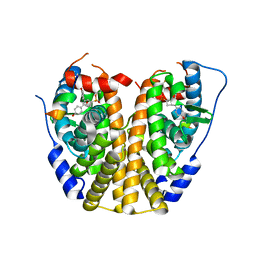 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Fluoro-substituted OBHS derivative | | Descriptor: | 3-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNU
 
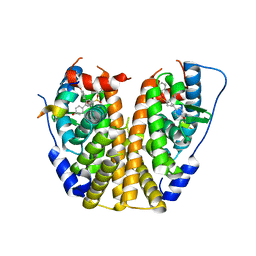 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methyl-substituted OBHS derivative | | Descriptor: | 2-methylphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNW
 
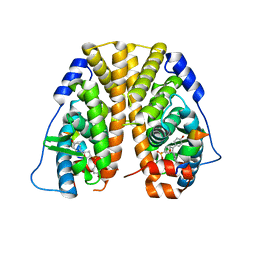 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 4-Bromo-substituted OBHS derivative | | Descriptor: | 4-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNH
 
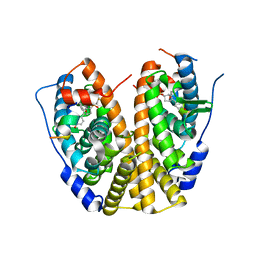 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Fluoro-substituted OBHS derivative | | Descriptor: | 2-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Bromo-substituted OBHS derivative | | Descriptor: | 3-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
