7ZLY
 
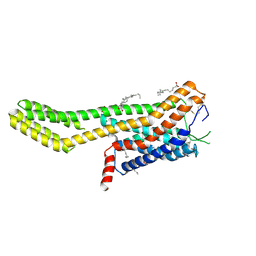 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
1KAK
 
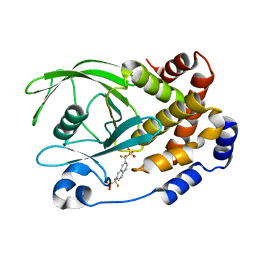 | | Human Tyrosine Phosphatase 1B Complexed with an Inhibitor | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, {[7-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALEN-2-YL]-DIFLUORO-METHYL}-PHOSPHONIC ACID | | Authors: | Jia, Z, Ye, Q, Dinaut, A.N, Wang, Q, Waddleton, D, Payette, P, Ramachandran, C, Kennedy, B, Hum, G, Taylor, S.D. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
J.Med.Chem., 44, 2001
|
|
4IGP
 
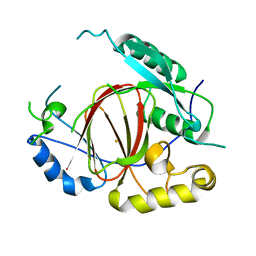 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 apo enzyme | | Descriptor: | FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGO
 
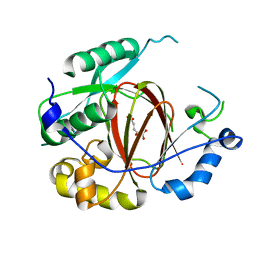 | | Histone H3 Lysine 4 Demethylating rice Rice JMJ703 in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGQ
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 in complex with methylated H3K4 substrate | | Descriptor: | FE (III) ION, N-OXALYLGLYCINE, Os05g0196500 protein, ... | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IJS
 
 | |
8XVV
 
 | | The TRRAP module of human NuA4/TIP60 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Isoform 2 of E1A-binding protein p400, Isoform 2 of Transformation/transcription domain-associated protein | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex
Nat Commun, 2024
|
|
8XVT
 
 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex
Nat Commun, 2024
|
|
8XVG
 
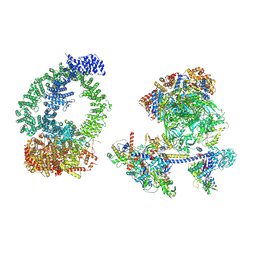 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex
Nat Commun, 2024
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
6IQT
 
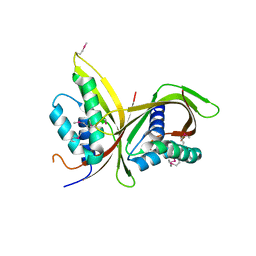 | | Crystal Structure of CagV, a VirB8 homolog of T4SS from Helicobacter pylori Strain 26695 | | Descriptor: | Cag pathogenicity island protein (Cag10) | | Authors: | Wu, X, Zhao, Y, Sun, L, Jiang, M, Wang, Q, Wang, Q, Yang, W, Wu, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-13 | | Last modified: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Crystal structure of CagV, the Helicobacter pylori homologue of the T4SS protein VirB8.
Febs J., 286, 2019
|
|
4R3D
 
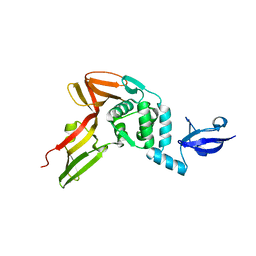 | | Crystal structure of MERS Coronavirus papain like protease | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Kong, L.Y, Wang, Q, Ming, Z.H, Shaw, N, Sun, Y.N, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | The Crystal Structures of Middle East Respiratory Syndrome Coronavirus Papain-like Protease and Its Complex with an Inhibitor
To be Published
|
|
6IZT
 
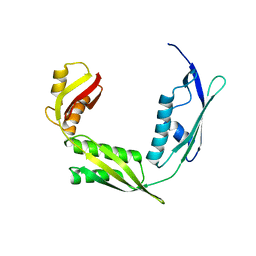 | | Crystal structure of Haemophilus Influenzae BamA POTRA3-5 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IZS
 
 | | Crystal structure of Haemophilus influenzae BamA POTRA4 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6VJT
 
 | |
1KAV
 
 | | Human Tyrosine Phosphatase 1B Complexed with an Inhibitor | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, [(4-{4-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-BUTYL}-PHENYL)-DIFLUORO-METHYL]-PHOSPHONIC ACID | | Authors: | Jia, Z, Ye, Q, Dinaut, A.N, Wang, Q, Waddleton, D, Payette, P, Ramachandran, C, Kennedy, B, Hum, G, Taylor, S.D. | | Deposit date: | 2001-11-03 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
J.Med.Chem., 44, 2001
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
1M8T
 
 | | Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Phospholipase a2 | | Authors: | Xu, S, Gu, L, Wang, Q, Shu, Y, Lin, Z. | | Deposit date: | 2002-07-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6CXC
 
 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
6PZT
 
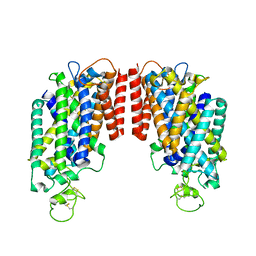 | | cryo-EM structure of human NKCC1 | | Descriptor: | Solute carrier family 12 member 2 | | Authors: | Cao, E, Wang, Q, Yang, X. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the human cation-chloride cotransporter NKCC1 determined by single-particle electron cryo-microscopy.
Nat Commun, 11, 2020
|
|
4RWT
 
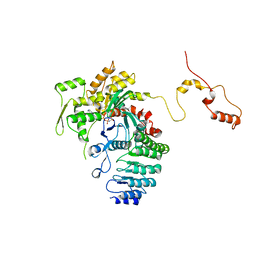 | | Structure of actin-Lmod complex | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2014-12-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
